A method for identification of radish clubroot resistance
A radish root and resistance technology, which is applied in the field of vegetable crop genetics and breeding, and achieves the effects of low equipment and technical requirements, good stability and product separation effect, and simple and convenient detection method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Identification of radish germplasm resistance to clubroot
[0033] The radish germplasm seeds were sterilized, rinsed and germinated, and then sown in a plug tray containing vermiculite: peat = 1:1 sterilization substrate. 6 seeds per accession, 1 per hole, were repeated 3 times, and protective rows were planted around the test material to prevent marginal effects.
[0034] After sowing, each hole was injected with 2 mL of 'XY-2' P. brassicae bacterial solution for inoculation (spore concentration 1×10 7 mL -1 ).
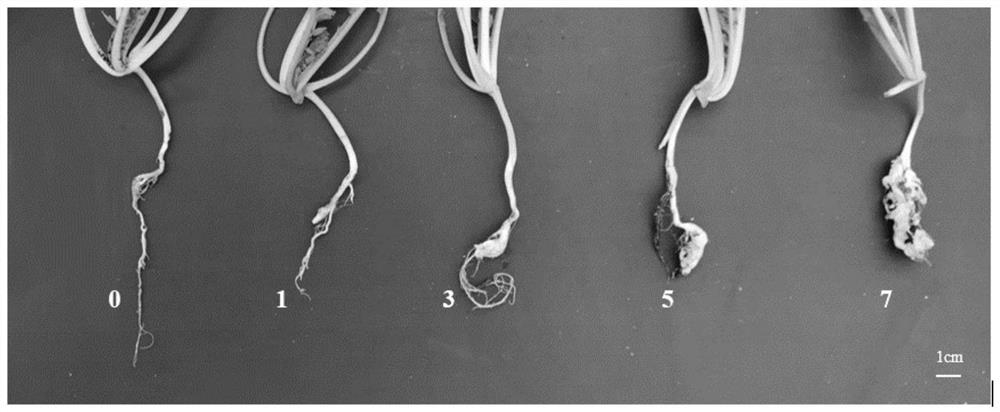
[0035] After 40 days of inoculation, the root disease of each plant tested was investigated, identified and analyzed, identified and graded one by one.
[0036] refer to as figure 1 The grading standard for the identification of radish clubroot disease at the seedling stage shown in: Grade 0, each root system grows normally, no gall mass; Grade 1, the main root does not swell, but there are small galls on the fibrous root; Grade 3, the main roo...
Embodiment 2
[0042] Example 2: Development and application of radish clubroot resistance markers based on RsNBS135 gene
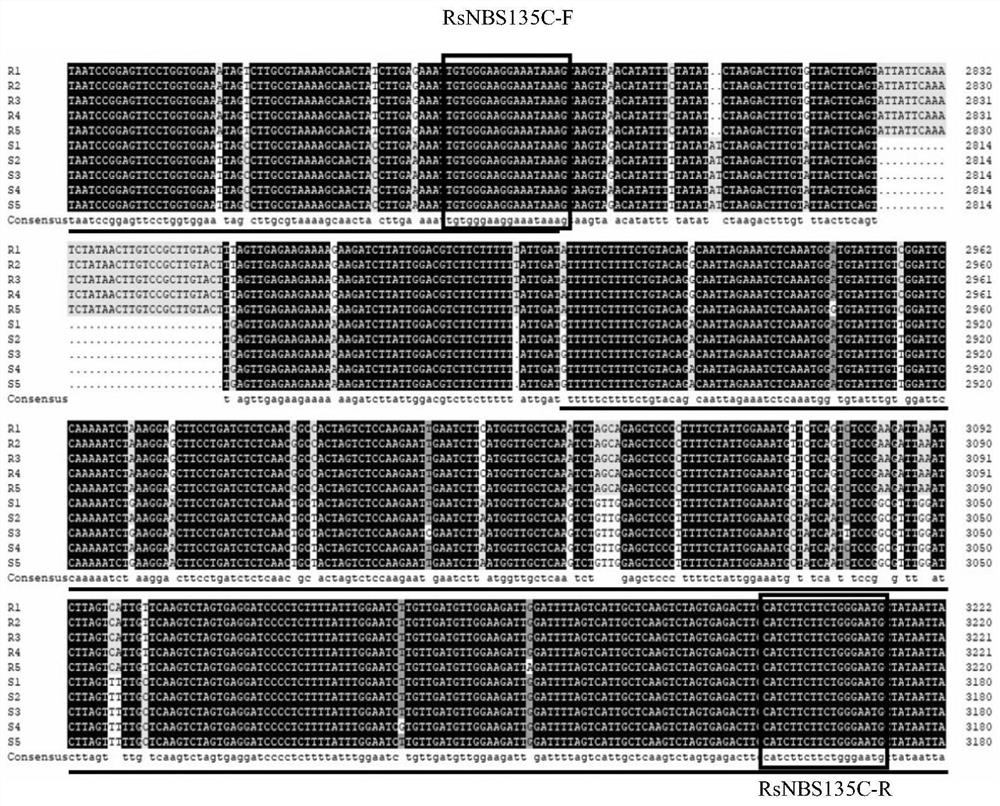
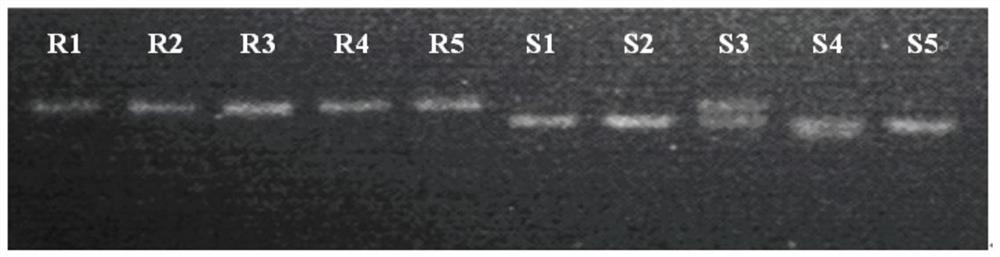
[0043] The radish whole genome nucleotide sequence, annotated CDS sequence and amino acid sequence were downloaded from the radish genome database (http: / / radish.kazusa.or.or.jp / ). The radish genome protein sequence was searched using the Markov Model (HMM, Hidden MarkovModel) according to the NBS protein domain (NB-ACR, PF00931) (http: / / pfam.xfam.org / family / PF00931), and the E-value was set as 0.01. All sequences were submitted to Pfam, NCBI and SMART to verify the sequence reliability. After removing the NBS-free and incomplete sequences, the identified 188 disease resistance candidate genes containing NBS structures were named RsNBS001-RsNBS188, respectively. The TIR-NBS-LRR gene RsNBS135 is closely related to the resistance of clubroot (race 4) through the sequence difference analysis of clubroot resistant germplasm. RT-qPCR was used to analyze the expression of RsN...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com