Rapid detection method for bacteroides based on high-throughput sequencing and application
A technology of Bacteroides and genes, applied in the field of molecular biology, to achieve accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1: Construction of Bacteroides-specific primers based on the high-resolution core gene of Bacteroides
[0036] 1. Through the database NCBI (National Center for Biotechnology Information) and EMBL
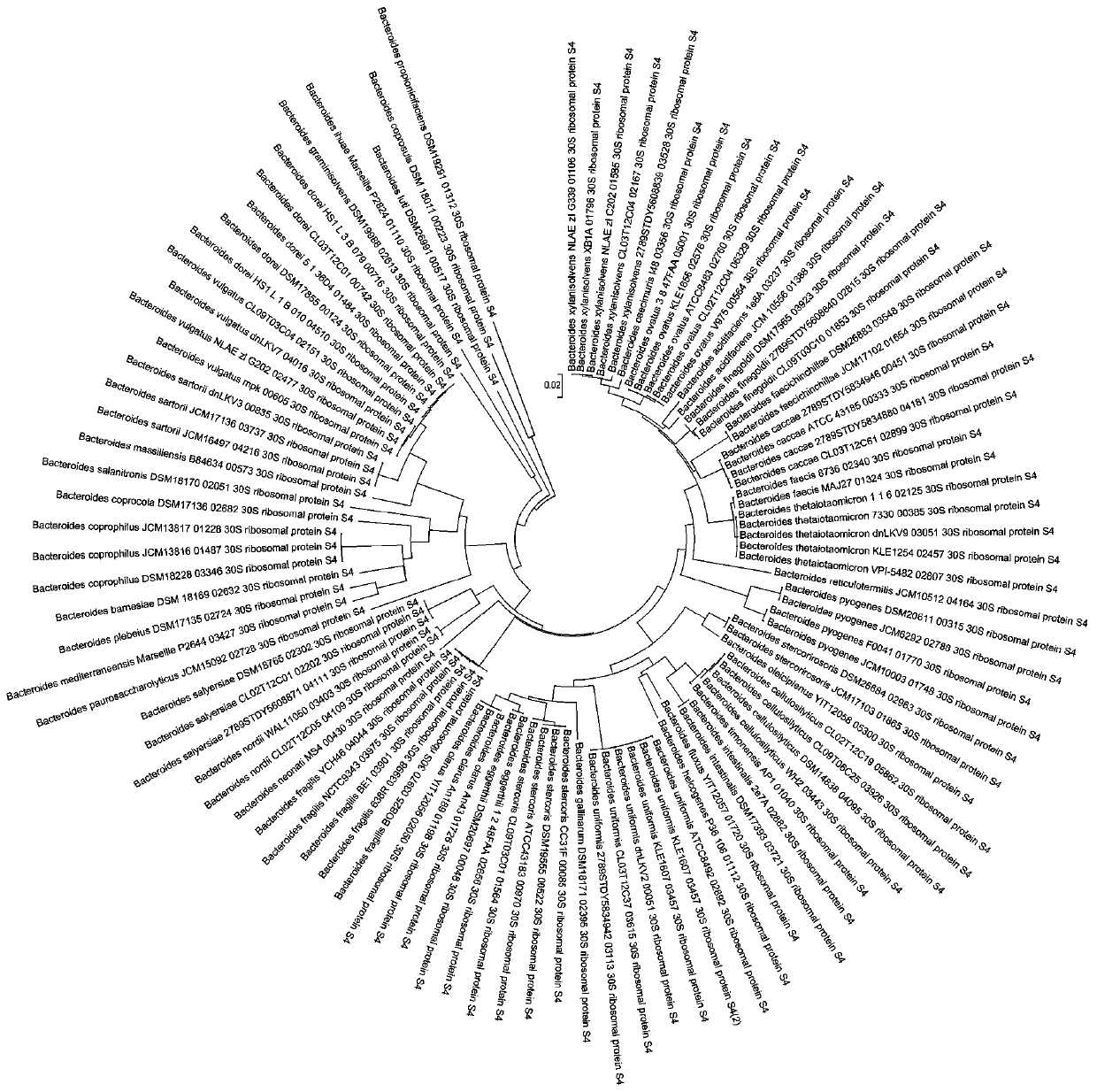
[0037] (European Molecular Biology Laboratory) research, 42 different Bacteroides species have completed genome sequencing, some have only one genome sequence, and some have nearly a hundred. We downloaded a total of 100 rpsD gene sequences of different species of Bacteroides (at least one sequence for each Bacteroides, and for those with more sequences, select 5 sequences with higher sequencing levels), and constructed a phylogenetic tree with MEGA software as follows: figure 1 As shown, we can see from the figure that this gene can well distinguish different species of Bacteroides.
[0038] 2. Construct the rpsD gene database, download the known rpsD gene sequences of Bacteroidetes from databases such as NCBI and EMBL, and use the downloaded sequences to construct...
Embodiment 2
[0049] Example 2: Validation of Bacteroides-specific primers at the genus level
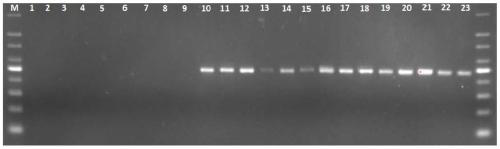
[0050] Bifidobacterium longum, Lactobacillus breris, Lactobacillus breris, Lactobacillus fermentum, Pediococcus acidilactici, Enterococcus faecalis, Escherichia coli ( Escherichia coli), Akkermansia muciniphila, Bacteroides caccae, Bacteroides dorei, Bacteroides eggerthii, Bacteroides faecis, Bacteroides salyersiae, Bacteroides uniformis, Bacteroides ovatus, Bacteroides fragilis, Bacteroides vulgatus, Bacteroides stercoris, Bacteroides xylanisolvens, Bacteroides thetaiotaomicron , Bacteroides kribbi, Bacteroides koreensis, respectively extract the genome, microbial genome extraction refers to the instructions in the bacterial genome DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd., Beijing, China), with sequences such as SEQ ID NO.1 and SEQ ID The primers shown in NO.2 are used for PCR amplification using the extracted genome as a template, and the amplification conditions ...
Embodiment 3
[0056] Example 3: Construction of a rapid detection method for Bacteroidetes and its verification at the species level
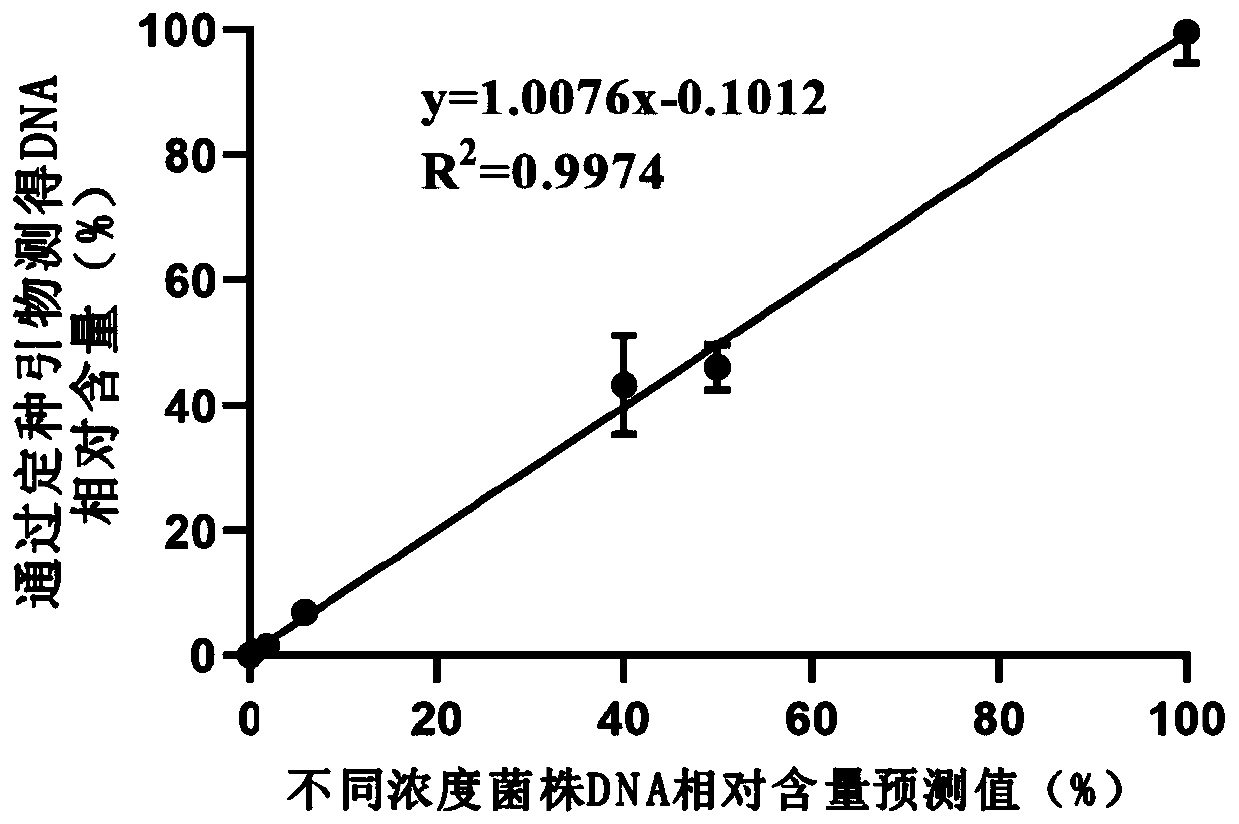
[0057]In order to verify the accuracy of primers SEQ ID NO.1 and SEQ ID NO.2 in the analysis of Bacteroides, we will common Bacteroides (Bacteroides vulgatus), Bacteroides salyersiae, Bacteroides dorei, Bacteroides faecis, Bacteroides uniformis (Bacteroides uniformis) , Bacteroidescaccae, Bacteroides; xylanisolvens, Bacteroides nordii, Bacteroidesstercoris, Bacteroides fragilis, a total of 10 species of Bacteroides (as shown in Table 2) genome according to the concentration of 0.001-50ng / μL Mixed, for the detection of different species of Bacteroides in simulated complex samples. Based on the newly designed Bacteroides identification primers combined with a next-generation sequencer, then perform analog sample sequencing according to the method steps described in Example 2, and finally compare the rpsD gene sequence information obtained by sequencing with th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com