Sample preparation method for proteomic analysis
A proteomics, protein technology, applied in the field of proteomics analysis, can solve the problem of insufficient recovery liquid-mass spectrometry analysis and other problems, achieve the effect of reducing the chance of sample contamination, improving the identification and quantification effect, and widening the scope of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
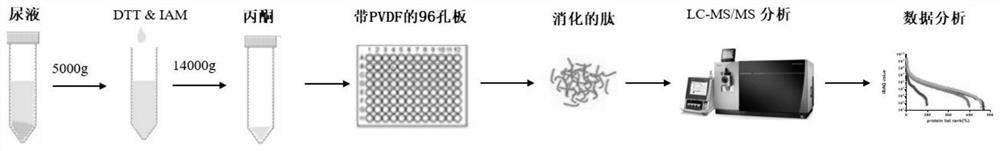
[0035] Specifically, the present invention relates to a sample preparation method for proteomics analysis, the method includes reducing and alkylating the protein, then adding pre-cooled acetone at -20°C for 30 minutes to precipitate, and the precipitate After the protein is reconstituted, it is transferred to the activated PVDF membrane pre-placed in a 96-well plate, so that the protein is adsorbed on the PVDF membrane, and the protein is rinsed and rinsed with tris solution on the membrane by using a suction filtration device. Replacement, followed by adding trypsin for enzymatic hydrolysis on the membrane, and finally the peptide solution on the membrane was eluted with acetonitrile methanol water solution, and the peptide was reconstituted for mass spectrometry analysis after draining. This sample processing method combines the steps of protein purification and 96-well plate membrane enzyme digestion. The purification step before protein membrane enzyme digestion provides a...
Embodiment 1
[0039] Example 1. Collection of protein-containing samples
[0040] Twelve normal subjects (6 males, 6 females) from Peking Union Medical College volunteers mixed morning urine. Urine samples were centrifuged at 4500 × g for 10 min at 4°C to remove cell debris, and the supernatant was aliquoted for subsequent analysis.
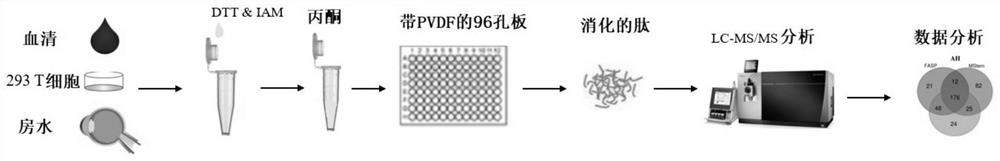
[0041] 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM, 4.5g / L glucose, 10% FCS, 2mM L-glutamine, 50mg / mL penicillin, 50mg / mL streptomycin) until the cell confluence was about 90%. Wash 3 times with 5 mL of ice-cold PBS. Cells were dissolved in 50 mM Tris-HCl with 8M urea, 1% NP-40, 5% SDS, and then incubated at 96°C for 5 minutes, followed by six 30s ultrasonic cycles (Bioruptor Pico, Diagenode, USA). The urea lysate was incubated at room temperature for 10 min, sonicated, and the supernatant protein solution after centrifugation was used for subsequent analysis.
[0042] The blood was centrifuged at 4000×g for 5 minutes, and the superna...
Embodiment 2
[0045] Example 2. Protein digestion
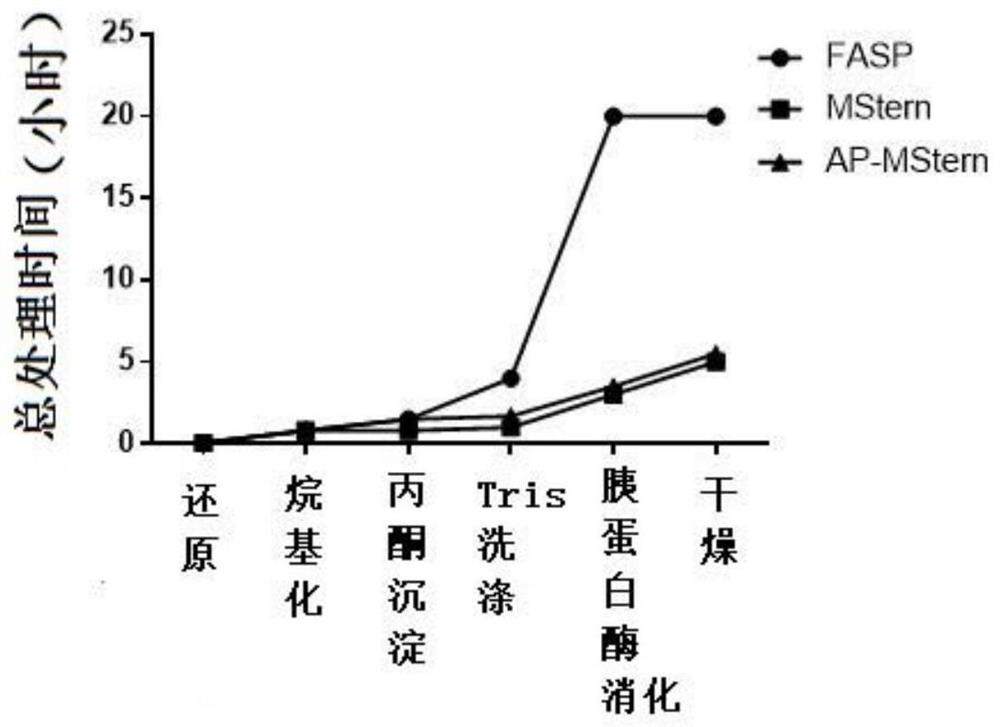
[0046] FASP method
[0047] Digestion was carried out according to the FASP method reported in the literature, see Wisniewski, J.R. et al., Universal sample preparation method for proteome analysis. Nat Methods, 2009.6(5): p.359-62. Specifically, the reducing agent DTT was added to the sample containing approximately 100 micrograms of protein (obtained in Example 1), and reacted in a water bath at 95°C for 5 minutes; after cooling, the alkylating agent IAM was added, and dark treatment was performed for 45 minutes. Add pre-cooled acetone 6 times the volume of the sample, vortex and mix well, put it in a -20°C refrigerator for half an hour, and then centrifuge at 12000g at 4°C for 10 minutes to obtain a protein precipitate. The protein was reconstituted with dissolving solution (2% SDS, 20mM Tris), transferred to a pre-washed 30KD ultrafiltration tube for centrifugation, and washed 3 times with 20mM Tris. According to the protein: enzyme ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com