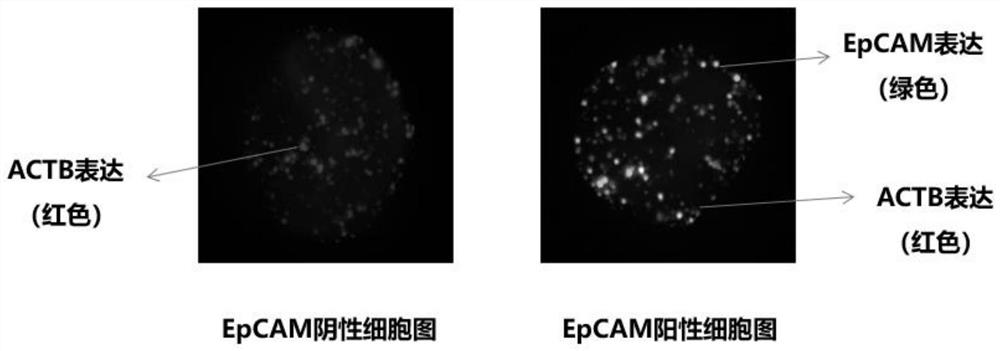
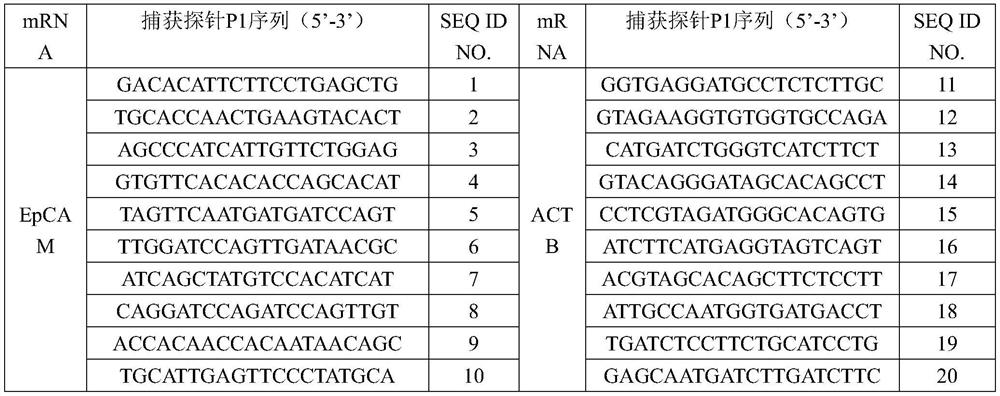
EpCAM gene expression detection kit
A detection kit and gene expression technology, applied in the field of molecular biology, can solve the problem of difficult control of factors such as limited sample sources and the accuracy of test results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 EpCAM gene expression detection kit
[0049] The EpCAM gene expression detection kit (in situ hybridization method) described in the present embodiment mainly includes:
[0050] 1. Capture Probes
[0051] From the 5' end to the 3' end of the capture probe is a specific P1 sequence capable of binding to the target mRNA to be detected, a solubilizing group, and a P2 sequence capable of binding to the P3 sequence of the amplification probe, aiming at the same target mRNA The P2 sequence in the capture probe is the same; the specific P1 sequence is a peptide nucleic acid sequence with a base length of 16-20bp; the solubilizing group is selected from O-linker, E-linker, X-linker At least one: the P2 sequence is a sequence that does not have a hairpin structure, does not form dimers or mismatches within and between probes, and does not specifically bind to P1 and EpCAM gene mRNA. The solubilizing group can improve the solubility of the capture probe, and at the sa...
Embodiment 2
[0077] Example 2 Using the kit described in Example 1 to detect the sample
[0078] The formula of described various solutions is as table 6:
[0079] Table 6
[0080]
[0081] In this embodiment, the blood samples of tumor patients are preferred, and the expression level of EpCAM gene of circulating tumor cells in the samples is detected, wherein the capture probe mixture, the amplification probe mixture, and the chromogenic probe mixture all use the reagents described in Example 1 All probes in the box corresponding to the list.
[0082] 1. Extract 5ml of blood from the patient's vein into a vacuum blood collection tube to obtain a blood sample
[0083] 2. Sample pre-treatment, the cells to be tested are filtered onto the filter membrane
[0084] (1) Transfer 5ml of blood sample to a centrifuge tube containing 10ml of erythrocyte lysate, shake and mix well for 10 seconds, centrifuge at 600×g for 5 minutes, discard the supernatant to keep the cell pellet, and resuspend ...
Embodiment 3
[0132] Embodiment 3 The impact of different types of capture probes on the detection effect of the kit
[0133] In order to evaluate the detection effect of kits composed of different types of capture probes, experimental groups 1-2 were designed. Except for the different types of capture probes, the other components of the two groups were the same. The specific design is shown in Table 9.
[0134] Table 9 Selection of kit capture probes
[0135] test group capture probe type Experimental group 1 Capture probes of the present invention Experimental group 2 Traditional Linear Oligonucleotide Probes
[0136] Using the kit designed and prepared above, the blood samples of 10 tumor patients (numbered 1-10) were detected according to the detection process and method described in Example 2, and the cells with DAPI blue fluorescent signal in each sample were read. , count the number of cells expressing green / red fluorescence, and the cells expressing t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com