Method for detecting molecular markers based on KASP technology
A technology of molecular markers and technical detection, which is applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as complex process, long time-consuming, error-prone, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
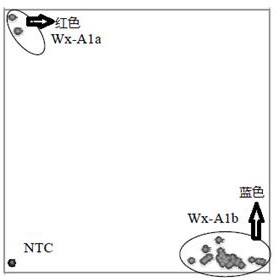
[0029] Wheat waxy gene has been reported Wx-A1 位于7A染色体,其分子标记MAG264为共显性STS标记,在野生型264A1a的扩增片段如SEQ No.3所示;具体序列为:CGGCGGTGCCTCTCCATGGTGGTGCGCGCCACGGGCAGCGGCGGCATGAACCTCGTGTTCGTCGGCGCCGAGATGGCGCCCTGGAGCAAGACTGGCGGCCTCGGCGACGTCCTCGGGGGCCTCCCCGCCGCCATGGCCGTAAGCTTGCGCCACTGCCTTCTTATAAATGTTTCTTCCTGCAGCCATGCCTGCCGTTACAACGGGTGCCGTGTCCGTGCAGGCCAACGGTCACCGGGTCATGGTCATCTCCCCGCGCTACGACCAGTACAAGGACGCCTGGGACA
[0030] 在突变体264A1b的扩增片段如SEQ No.4所示;具体序列为:CGGCGGTGCCTCTCCATGGTGGTGCGCGCCACGGGCAGCGGCGGCATGAACCTCGTGTTCGTCGGCGCCGAGATGGCGCCCTGGAGCAAGACTGGCGGCCTCGGCGACGTCCTCGGGGGCCTCCCCGCCGCGGACGCCTTCTTATAAATGTTTCTTCCTGCAGCCATGCCTGCCGTTACAACGGGTGCCGTGTCCGTGCAGGCCAACGGTCACCGGGTCATGGTCATCTCCCCGCGCTACGACCAGTACAAGGACGCCTGGGACA
[0031] Using the 264A1a sequence to blast the wheat reference genome on the URGI website, a highly similar sequence was found at 7D and 4A.
[0032] After downloading similar sequences, DNAMAN software was used for multiple sequence alignment, and it was determined that the specific SN...
Embodiment 2
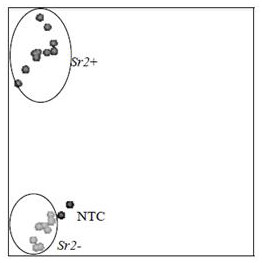
[0041] Wheat stem rust resistance gene has been reported Sr2 Located on chromosome 3B, the CAPS molecular marker csSr2 is dominant in most wheat materials in my country, similar to the molecular marker SCAR. The csSr2 amplified fragment DNA sequence is shown in SEQ No.8; specifically:
[0042] CAAGGGTTGCTAGGATTGGAAAACAAAATTGTATTCTTTGTGATTGTAATTGTTTGGTTCAAACATGAATAAAATAATATTCGATTTTCCATTGCTCTACACAACAAGTGTTCATGATTCTAGAACCCTCAAATGGTCGAGCACAAGCTCTAATTTCTTTGGAATCATGAGTGTTGGATGTCTCGCACACCTAGCTTAATACATGCAGATACTTGTTGGGAAGTAACGCTGCTAAAGAAACTTATGTATGGTTCTTAGCTAAATTAGGTTTGAGAGGGGCTAATGGTTCTATGTTGTGTAGCTTGCCCAGAAAAATGTAAGATCATAAGAGTTATCT
[0043] Compare it with the reference genome by BLAST, find multiple high-similarity sequences on the 3D and 3B chromosomes, and use DNAMAN for visual comparison after downloading. Design a site-specific primer Sr2H3 at a known SNP site, as shown in SEQ No.9 (5'-CGAGCACAAGCTCTAATTTCTTTGG AATCA-3'), the Tm value is 59 degrees, another SNP site is found at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com