A kind of primer set, kit and application thereof for sars-cov-2 virus nucleic acid detection
A sars-cov-2 and kit technology, applied in the field of biotechnology applications, can solve the problems of limited detection throughput and high price, and achieve the effect of good repeatability, strong specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0070] 3. Preparation of guide DNA and molecular beacons
[0071] (1) Preparation of guide DNA:
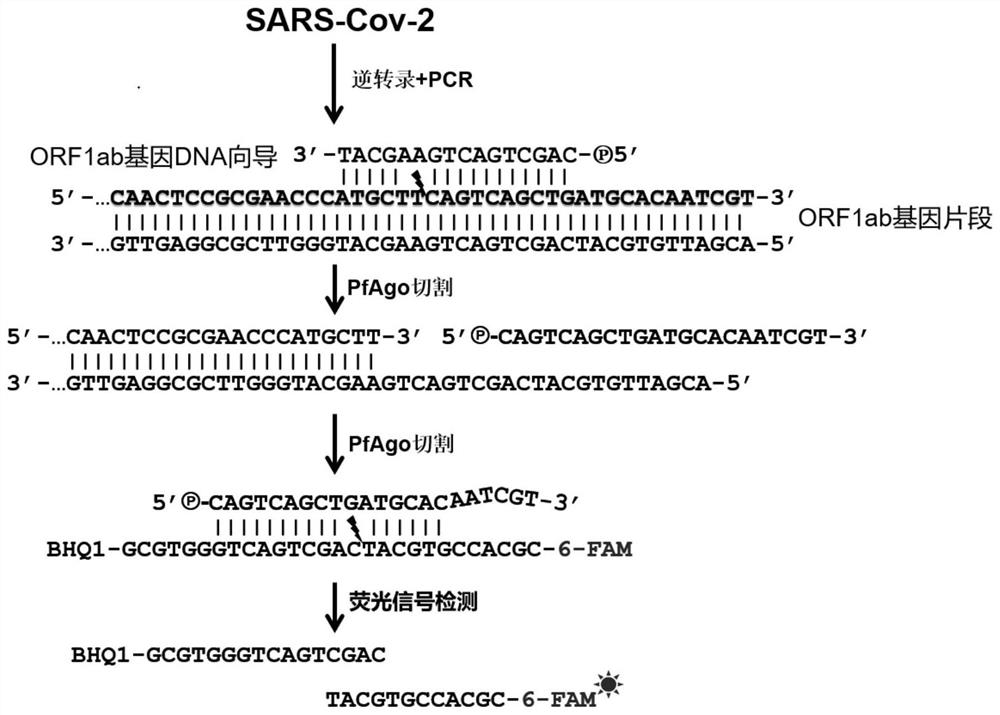
[0072] The guide DNA guides the PfAgo enzyme to cleave the target DNA. All the guide DNAs in the examples of the present invention were treated with T4 polynucleotide kinase (T4 PNK) to make the 5' end with a phosphate group. The T4 PNK reaction system is shown in Table 1.
[0073] Table 1 Phosphorylation modification reaction system configuration of guide DNA
[0074]
[0075] (2) Preparation of molecular beacons:
[0076] In this embodiment, the 5' end and the 3' end of the molecular beacon are respectively modified by a fluorescent group and a quencher group, and the fluorescent group is selected from at least one of FAM, VIC, Cy5 or Rox; the The fluorescence quenching group is selected from at least one of BHQ-1, BHQ2, BHQ-3, BBQ or TAMRA. In other embodiments, all other known fluorophores and fluorescence quenching groups can also be used for labeling. The specific exc...
Embodiment 2
[0083] The method for identifying or assisting in identifying whether a sample to be tested contains novel coronavirus SARS-CoV-2 provided by the present invention comprises the following steps:
[0084] S1. Using the nucleic acid extract in the sample to be tested as a template, add separate N gene primers, ORF1ab gene primers, or add N gene primers and ORF1ab gene primers at the same time for reverse transcription to obtain the cDNA of the new coronavirus SARS-CoV-2, The reverse transcription reaction system is shown in Table 4.
[0085] Table 4
[0086]
[0087] The reverse transcription reaction program is: 55°C, 15min; 85°C, 20min.
[0088]S2, using the above primer set, the cDNA containing a single target sequence (N gene or ORF1ab gene) obtained by the reverse transcription, and the cDNA containing both the N gene and the ORF1ab gene as templates, respectively performing single-plex PCR and double-PCR;
[0089] S3, mixing the guide DNA, molecular beacon and PfAgo e...
Embodiment 3
[0111] The present invention is used to identify or assist in the identification of the lower limit of detection of the detection method of the novel coronavirus SARS-CoV-2, comprising the following steps:
[0112] 1: Dilute the positive standard containing the N gene to 1000copies / μL, 100copies / μL, 10copies / μL, 1copies / μL, 0copies / μL;
[0113] 2: Carry out PCR amplification to the positive standard substance of the different concentration obtained by step 1 dilution respectively, PCR amplification method is the same as embodiment 2;
[0114] 3: The PCR product amplified in step 2 is subjected to a PfAgo digestion reaction, and the PfAgo digestion reaction is the same as in Example 2;
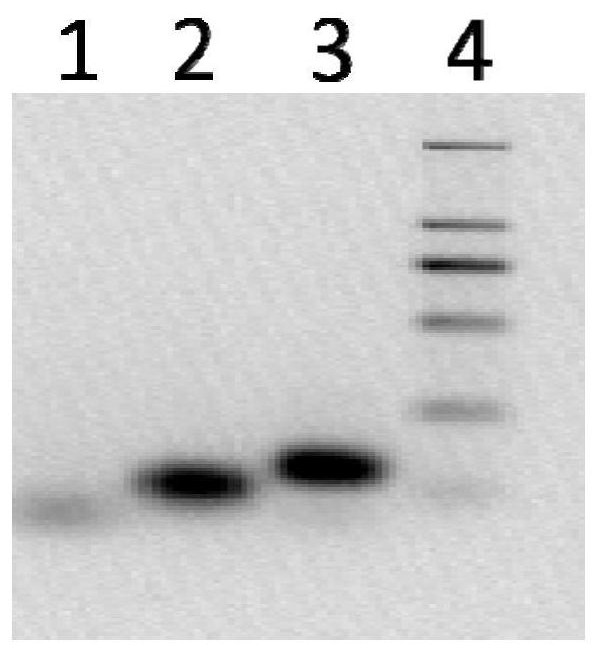
[0115] The amplified products obtained by PCR were subjected to agarose gel electrophoresis, and the results were as follows: Figure 7 Shown; For the detection of fluorescence value of PfAgo digestion, such as Figure 8 shown. The results show that the fluorescence detection values of eac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com