Prediction method for identifying 4-methylcytosine site
A methylcytosine and prediction method technology, which is applied in the field of prediction methods and software systems for identifying 4-methylcytosine sites, can solve the problems of lack of prediction algorithms, laborious and laborious problems, and achieves great application significance and excellent accuracy. , the effect of accurate prediction and identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
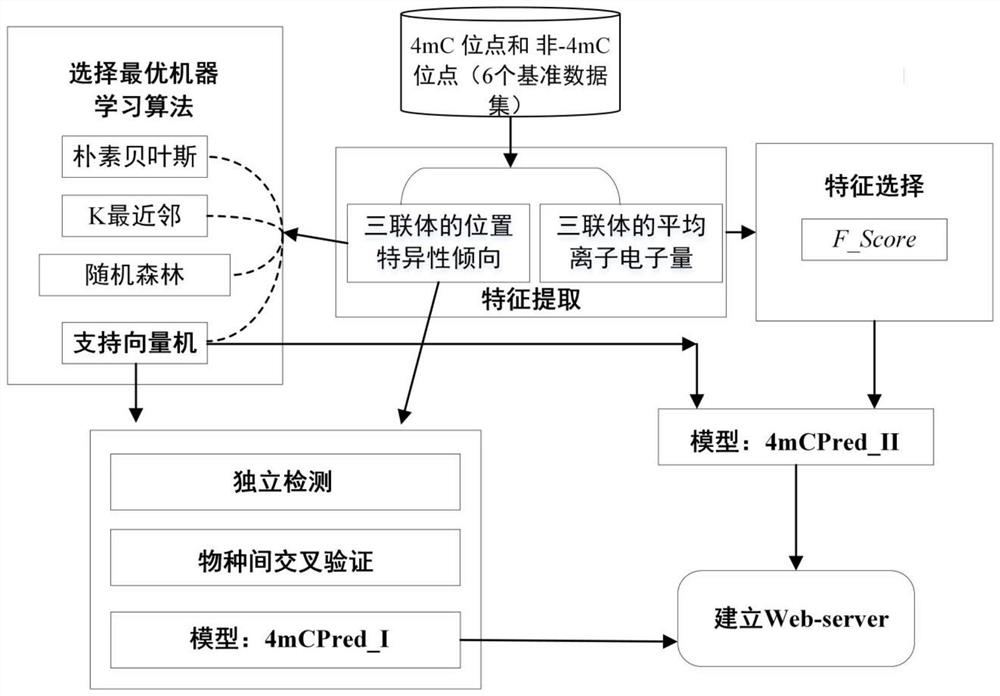
[0071] The 4-methylcytosine site is a specific methyltransferase (DNA methyltransferase, DNMT) that transfers a methyl group to the N4 position of cytosine to form a modification site. figure 1 It is a flowchart of a prediction algorithm for identifying 4-methylcytosine sites, and the specific steps are as follows:
[0072] Step 1: Establishment of benchmark dataset
[0073] Select Chen W, Yang H, Feng P, et al.iDNA4mC: identifying DNA N4-methylcytosine sites based on nucleotide chemical properties[J].Bioinformatics,2017,33(22):3518-3523Caenorhabditiselegans , Drosophila melanogaster, Arabidopsis thaliana, Escherichia coli, Geoalkalibacter subterraneus, Fusobacterium Geobacterpickeringii positive samples with 4-methylcytosine sites and without 4-methylcytosine sites Six benchmark datasets constructed from positive and negative samples of cytosine sites. In each species, the positive sample is a sequence fragment centered on 4-methylcytosine, with 20 bp upstream and downstrea...
Embodiment 2
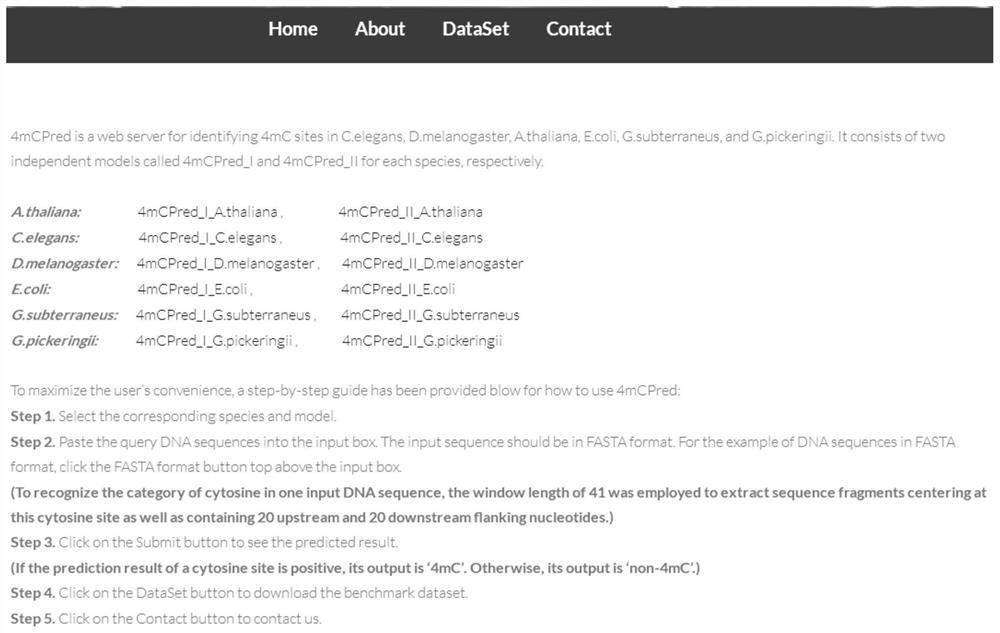
[0102] This embodiment provides a software system 4mCPred developed by a prediction algorithm for identifying 4-methylcytosine sites. Each species corresponds to two independent models 4mCPred_I and 4mCPred_II. Choosing 4mCPred_II can get the best prediction results, and choosing 4mCPred_I can better understand the position-specific tendency of triplets. The software system is a software system developed based on the optimal model using MATLAB software and JavaScript programming language. Users submit at least one DNA sequence in FASTA format, and they can quickly predict whether cytosine (C) in this sequence may be methylated (the length of the upstream and downstream sequences of this cytosine should not be less than 20).
[0103] To predict the possibility of methylation of cytosine in the DNA sequence of six species, the user only needs to select the model of the corresponding species in the prediction interface of 4mCPred, and input the FASTA format sequence of the corres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com