Glucose transporters and their use in enhancing organic acid production
A technology of glucose transport and protein, applied in the field of bioengineering and genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] 1. Construction of Glucose Transporter Expression Plasmid
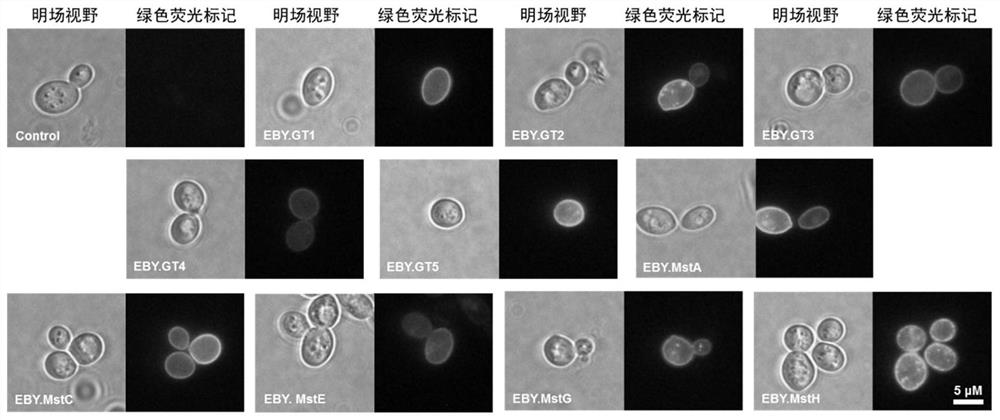
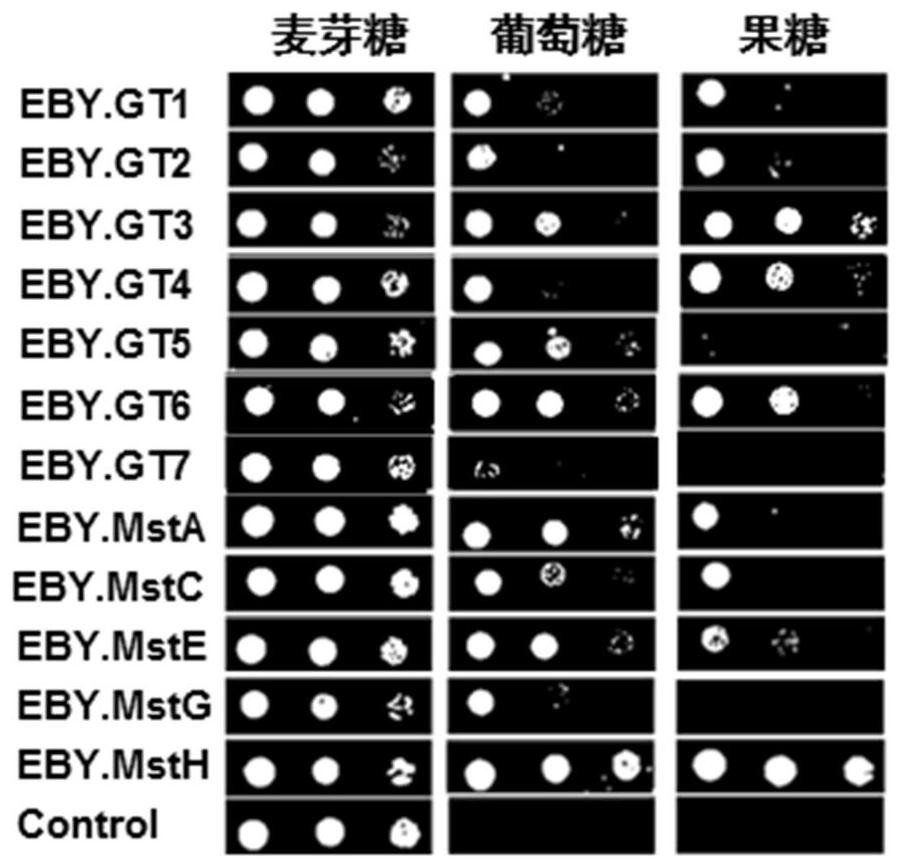
[0042] Based on genome data mining combined with transcriptome data analysis, seven potential glucose transporters GT1, GT2, GT3, GT4, GT5, GT6, GT7 were predicted, the amino acid sequences of which are shown in SEQ ID NO:1-7. Using the upstream primers and downstream primers of each glucose transporter, the cDNA of the citric acid producing strain D strain (purchased from the strain resource bank of Shanghai Industrial Microbiology Technology Co., Ltd. (formerly Shanghai Institute of Industrial Microbiology), public preservation number M202) was Templates were used to amplify the cDNA fragments of each glucose transporter by PCR. The specific primer sequences are shown in Table 1.
[0043] Table 1 Primers constructed by glucose transporter expression plasmids in Saccharomyces cerevisiae
[0044]
[0045]
[0046] The PCR reaction system was 10 μL of 5× FastPfu buffer, 1 μL of 10 mM dNTPs, 2.5 μL of upstr...
Embodiment 2
[0051] 1. Construction of glucose transporter Aspergillus niger expression plasmid
[0052] The upstream primers and downstream primers of each glucose transporter were used, and the genome of the citric acid producing strain D strain was used as a template to carry out PCR amplification of each glucose transporter gene fragment. The specific primer sequences are shown in Table 2. Although multiple glucose transporters have been reported in Aspergillus niger, the effect of their overexpression on citric acid fermentation is not clear. Therefore, in the present embodiment, 5 reported glucose transporters (MstA, MstC, MstE, MstG, MstH) are simultaneously analyzed, and their sequences are respectively SEQ ID NO: 8, SEQ ID NO: 9, SEQ ID NO: 10, SEQ ID NO: 11 and SEQ ID NO: 12 were overexpressed and detected by citric acid fermentation.
[0053] Table 2 The primers constructed by the glucose transporter Aspergillus niger expression plasmid
[0054]
[0055]
[0056] The PC...
Embodiment 3
[0074] 1. Construction of glucose transporter / citrate efflux protein Aspergillus niger expression plasmid
[0075] First, the upstream and downstream primers of citrate efflux protein CexA (the amino acid sequence of which is SEQ ID NO: 13) were used to amplify the citrate efflux protein gene fragment by PCR using the genome of citric acid producing strain D as a template. The specific primer sequences are shown in Table 5.
[0076] Table 5 The primers constructed by the glucose transporter Aspergillus niger expression plasmid
[0077]
[0078] The PCR reaction system was 10 μL of 5× FastPfu buffer, 1 μL of 10 mM dNTPs, 2.5 μL of upstream / downstream primers, 0.5 μL of DNA template, 1.5 μL of FastPfu (TransGene), and 32 μL of ultrapure water.
[0079] The PCR reaction conditions were: pre-denaturation at 95°C for 5 min; denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 2 min, 35 cycles; and final extension at 72°C for 10 min.
[0080] Aft...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com