Human iPS cell gene editing and screening method
A cell gene and screening method technology, applied in the field of gene editing and screening of human iPS cells, can solve the problems of expensive flow sorting instruments, high experimental conditions, and the unknown influence of human genome, etc., and achieve the convenience of single cloning Effects of selection and establishment of lines and improvement of gene editing rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
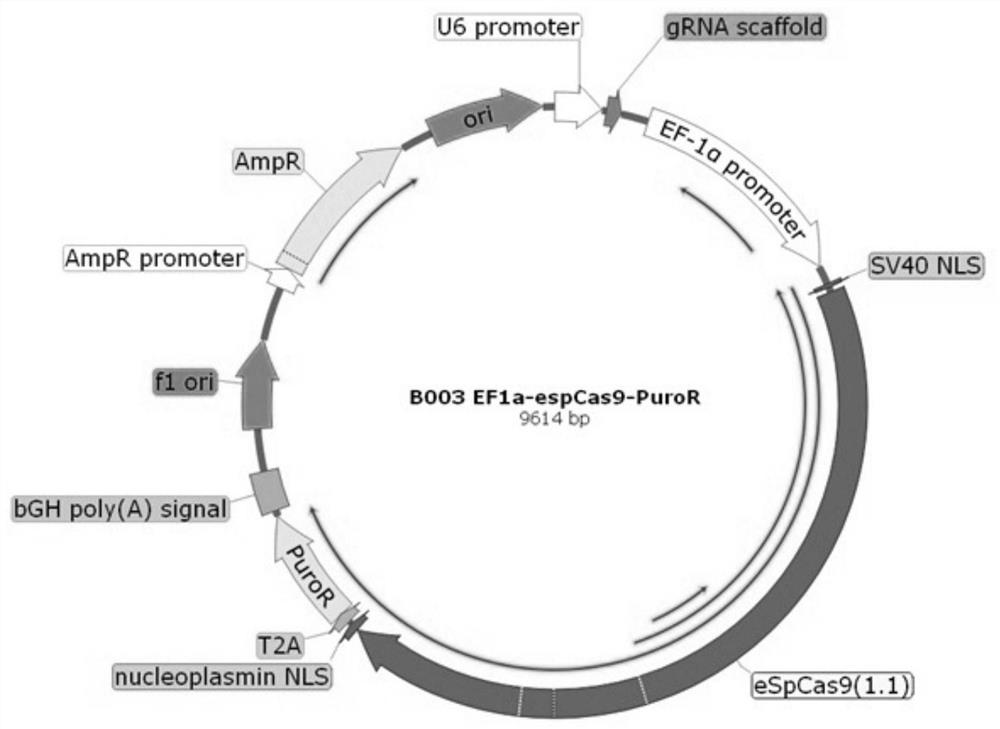
[0026] Example 1 is used for the construction of human iPS cell gene editing plasmid
[0027] An integrated plasmid (hU6-sgRNA-EF1a-eSPCas9(1.1)-2A-Puro ), the main frame of the plasmid is #64139 plasmid, eSPCas9(1.1) is derived from #71814 plasmid, and the EF1a promoter sequence is derived from #60599 plasmid. (The plasmids used were purchased from ADDGENE, the endonucleases were purchased from Thermo, and the one-step cloning kit was purchased from Novozyme)
[0028]1.1 was changed to eSPCas9 (1.1), and plasmid B002 was constructed
[0029] 1.1.1 #64139 plasmid linearization
[0030] 64139 plasmid 1ug Buffer R 2ul Bsm I 1ul EcoR V 1ul ddH2O Upto 20ul
[0031] Digest overnight at 37°C, inactivate at 80°C for 20 minutes
[0032] 1.1.2 Insert fragment PCR
[0033] #71814 plasmid 1ug EcoR I Buffer 2ul EcoR I 1ul ddH2O Upto 20ul
[0034] Digest overnight at 37°C, inactivate at 80°C for 20 minutes, t...
Embodiment 2
[0055] Example 2 Human iPS cell B2M gene knockout
[0056] Endonuclease and ligase were purchased from Thermo, mTeSR1, CloneR, ACCUTASE, and Metrigel were purchased from STEMCELL, and electroporation reagents were purchased from Lonza.
[0057] 2.1 Insert the B2M sgRNA sequence to construct the B2M knockout working plasmid (C003),
[0058] 2.1.1 sgRNA sequence primer synthesis: synthesize the following primers
[0059] B2Msg-F CACCGCGCGAGCACAGCTAAGGCCA seq ID No: 9 B2Msg-R AAACTGGCCTTAGCTGTGCTCGCGC seq ID No: 10
[0060] ddH2O dissolved primers to 10uM,
[0061] 2.1.2 B003 plasmid linearization
[0062] B003 plasmid 1ug FastDigest BpiI 1ul 10×FastDigest buffer 2ul ddH2O Upto 20ul
[0063] 37°C for 30 minutes
[0064] 2.1.3 Annealing dilution of sgRNA sequence
[0065] B2Msg-F (10uM) 4.5ul B2Msg-R (10uM) 4.5ul 10×PCR buffer 1ul
[0066] Put the PCR tube containing the mixture into a ...
Embodiment 3
[0085] Example 3 Editing Result Identification
[0086] 3.1 Cell line DNA extraction
[0087] When the iPS cells grow to cover 70% of the bottom of the well, aspirate the medium, add 1ml DPBS to each well to wash the residual medium, then aspirate and discard, add 1ml 0.5mM EDTA solution to each well, incubate at 37°C for 10 minutes, use a 1ml pipette Repeatedly blow and wash the bottom of the plate, transfer all the liquid to a 1.5ml centrifuge tube, centrifuge at 300g for 5 minutes, and remove the supernatant. Using Tiangen Whole Blood Genomic DNA Extraction Kit, follow the instructions to extract DNA.
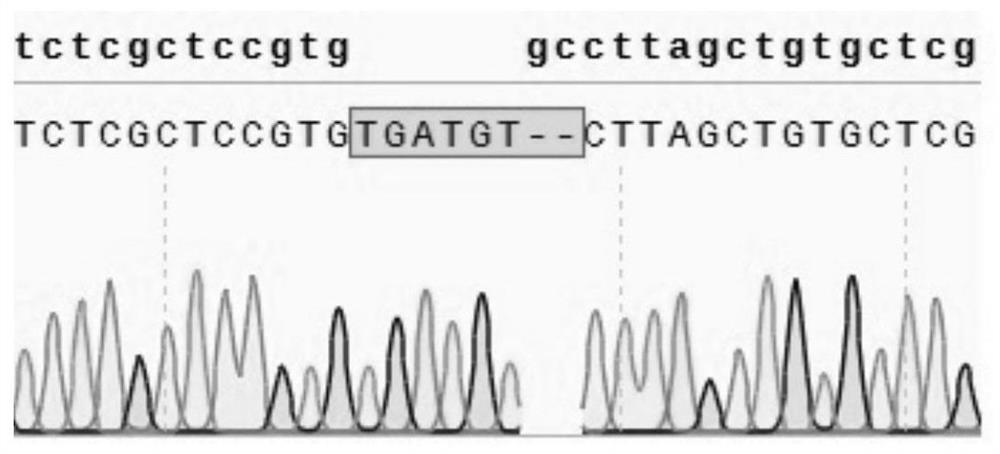
[0088] 3.2 Sequencing detection of editing results
[0089] Primers B2M-F: CAGCAAGGACATAGGGAGGAAC (seq ID No: 11) and B2M-R: CACCAAGGAGAACTTGGAGAAG (seq ID No: 12) were used for amplification, and the product was sequenced by Sanger method. The sequencing results of the cell line numbered EK33 are as follows: figure 2 As shown, the results show that the genome lost 2 ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com