A chemiluminescent biosensor for detecting 5-hydroxymethylcytosine, its detection method and application
A chemiluminescence detection and hydroxymethylcytosine technology, which is applied in the direction of chemiluminescence/bioluminescence, and analysis by making materials undergo chemical reactions, can solve the problems of non-specific amplification and false positives, and achieve high reproducibility and Accuracy, simple operation, and the effect of improving detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
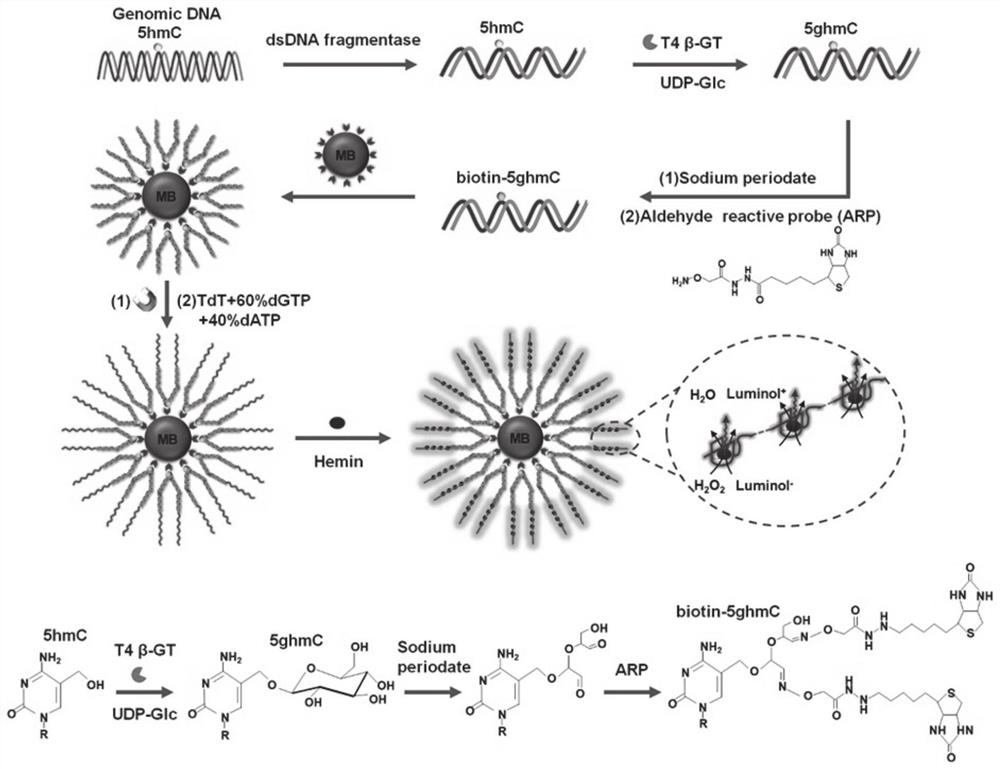
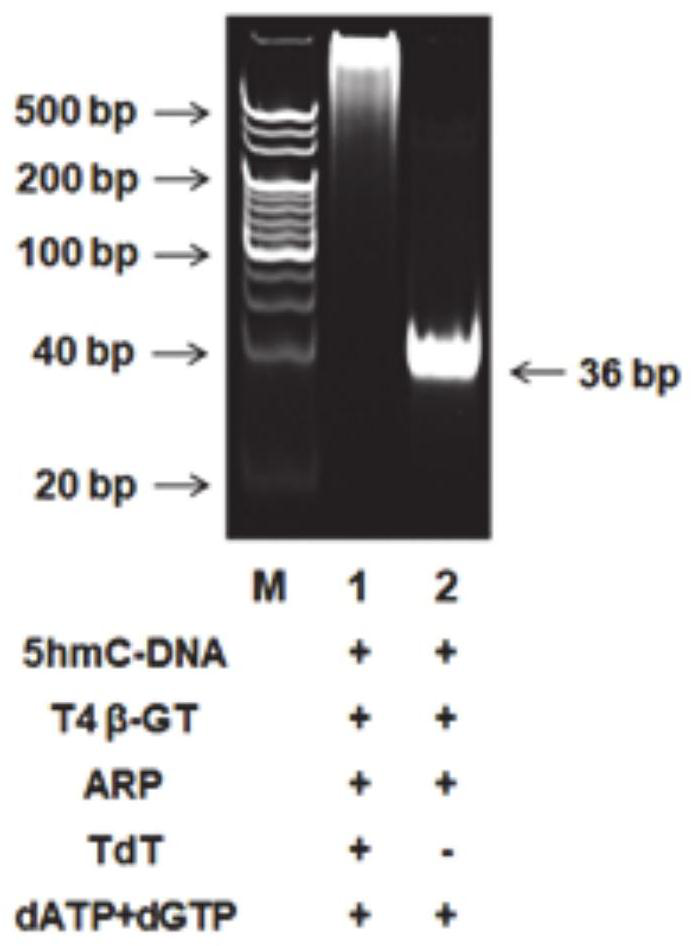
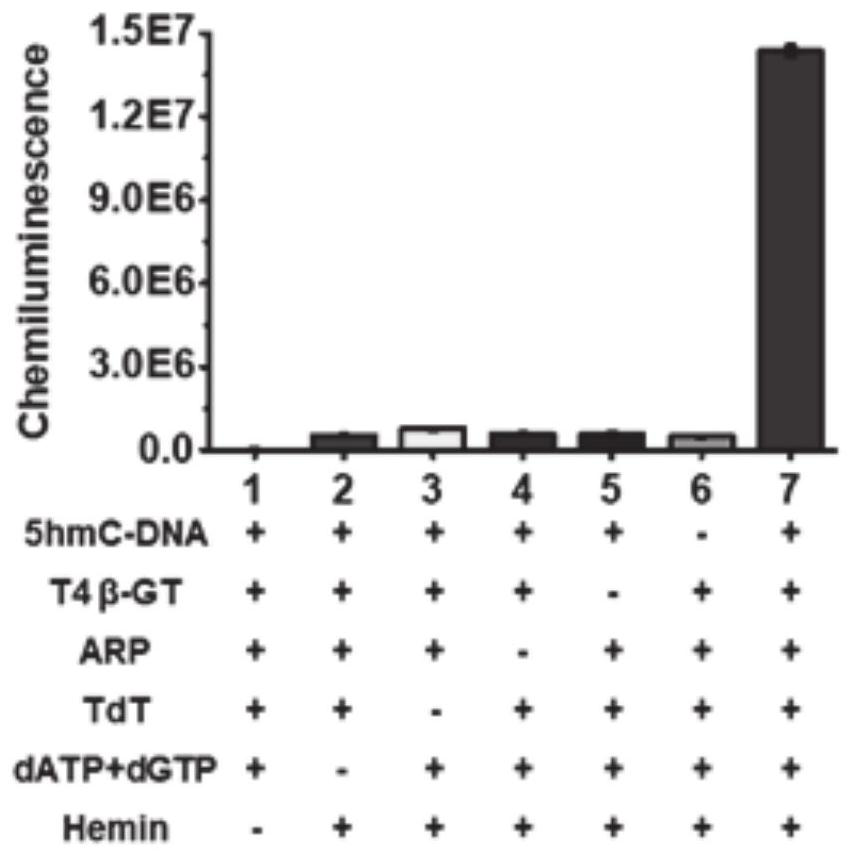
[0049] Preparation of biotinylated 5hmC-DNA products: The preparation of biotinylated 5hmC-DNA products involved three sequential steps, including glucosylation, iodic acid oxidation, and biotinylation. Perform the glucosylation reaction in 30 μl of reaction solution containing double-stranded DNA (dsDNAs), NEBuffer 4 (50 mM potassium acetate, 20 mM Tris-acetic acid, 10 mM magnesium acetate, 1 mM DTT, pH 7.9), 2 mM UDP-Glc and 4 U T4β-GT were reacted in a 37 °C water bath for 2 h, followed by a QIAquick nucleotide excision purification column to remove unreacted nucleotides and ions. The purified DNA was subjected to periodate oxidation in a 50 μl reaction system containing 20 mmol of sodium periodate and 100 mmol of sodium phosphate (pH 7.0) at 22°C for 16 h, and then reacted with 40 mmol of sodium periodate. Molar sodium sulfite was reacted at room temperature for 10 minutes, and then salt ions were removed using a QIAquick nucleotide excision cleanup column. The biotinylat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com