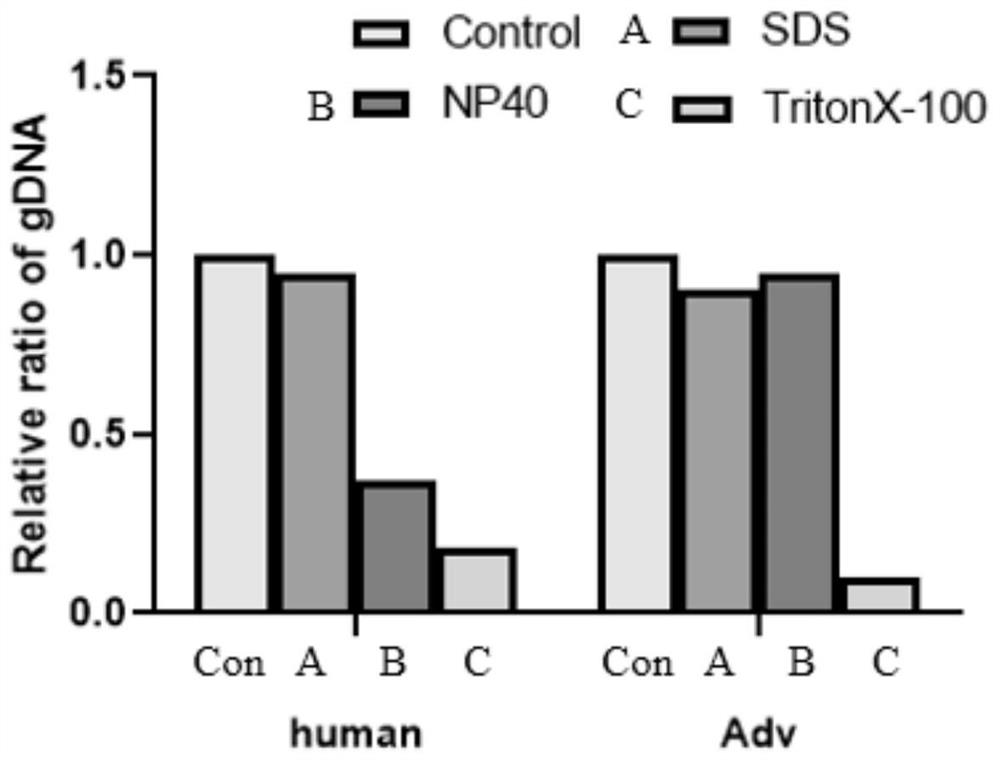
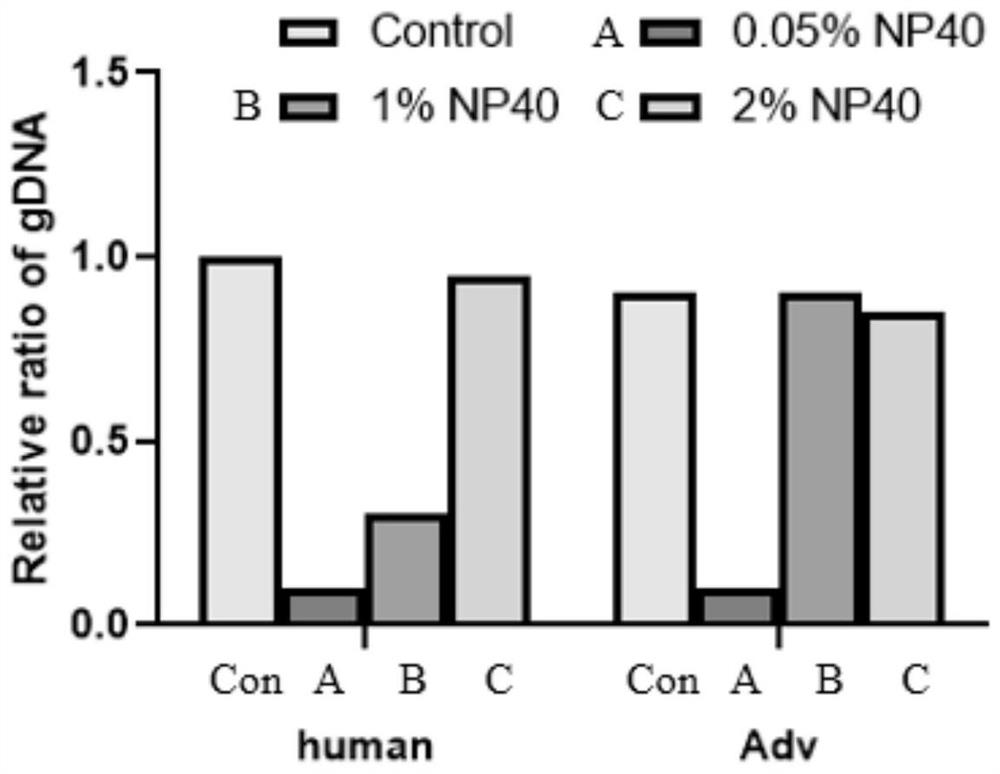
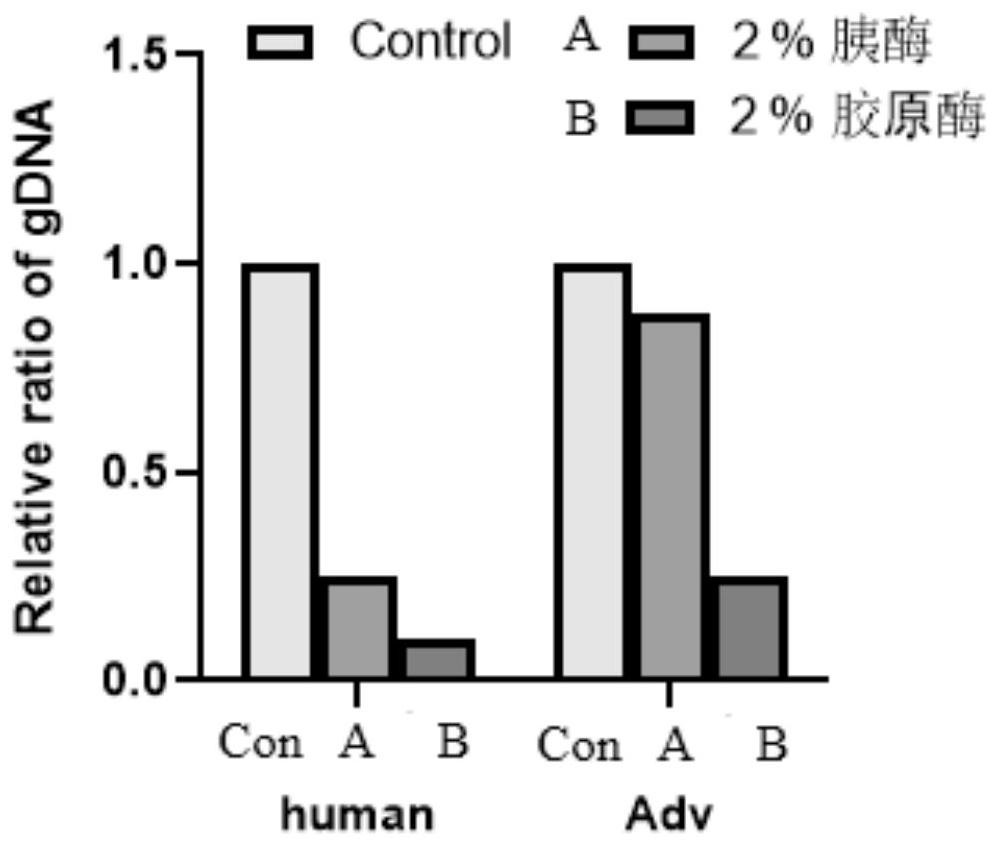
Intracellular microorganism enrichment method and reagent
A technology of microorganisms and cells, applied in biochemical equipment and methods, measurement/inspection of microorganisms, separation of microorganisms, etc., can solve problems such as improving sequencing depth and increasing sequencing costs, and achieve the effect of increasing the sequence ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] A method for enriching intracellular microorganisms, comprising the following steps:
[0047] 1. Collect cells
[0048] Cellular components of the sample are collected.
[0049] 1.1 Whole blood sample: Place the blood collection tube (containing whole blood, about 5-10ml) in a centrifuge, centrifuge at 1600g for 10min, transfer the upper layer of plasma, and use a pipette to absorb the middle layer of white blood cells.
[0050] 1.2 Other clinical infection samples containing cells: take 500-1000 μl of samples, centrifuge at 10,000-12,000 rpm for 5 minutes, and collect cells.
[0051] 2. Release microorganisms
[0052] Use cell treatment agents to make cell membranes more permeable and release intracellular microorganisms, specifically:
[0053] Take the cell fraction collected in step 1, and use 400-600 μl cell treatment agent to treat the cells (about 5x10 6 ), incubate on ice for 5-20 min, and shake and mix continuously during the period to increase the permeabil...
Embodiment 2
[0066] In this example, the method in Example 1 is used to detect whole blood samples.
[0067] 1. Sample preparation.
[0068] Divide the 5ml whole blood sample into 2 parts, one of which is centrifuged at 1600g for 10min, collect the upper layer plasma and the middle layer white blood cell samples into 2.0ml EP tubes, respectively marked as A and B, and the other whole blood sample Marked as C.
[0069] 2. Sample pretreatment and preparation.
[0070] Sample B (white blood cells) was treated with reference to steps 2-6 in Example 1; after sample A was directly extracted with the Micro DNA Extraction Kit (DP316) of Tiangen Biochemical Technology (Beijing) Co., Ltd., refer to Example 1 Steps 5-6 for processing; Sample C is processed with reference to steps 4-6 in Example 1. All three samples were constructed as qualified high-throughput (NGS) sequencing libraries.
[0071] 3. Detection
[0072] The A, B and C samples in the above step 2 were sequenced on the machine at th...
Embodiment 3
[0081] In this example, the method in Example 1 is used to detect the lower respiratory tract sample (alveolar lavage fluid).
[0082] 1. Sample preparation.
[0083] Divide 2ml of alveolar lavage fluid sample into 2 parts, one of which takes a certain volume (500-1000μl) sample, centrifuges at 10,000-12,000rpm for 5min, collects cells, and marks it as D; the other takes the same volume as sample D The original sample, labeled E.
[0084] 2. Sample pretreatment and preparation.
[0085] Sample D was processed with reference to steps 2-6 in Example 1; sample E was processed with reference to steps 4-6 in Example 1. Both samples were constructed into qualified high-throughput (NGS) sequencing libraries.
[0086] 3. Detection
[0087] The D and E samples in the above step 2 were sequenced on the machine at the same time, and the same sequencing depth was adopted, and the results were as follows:
[0088] Table 2 The sequencing results of the above D and E samples
[0089] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com