Kit for rapidly extracting gram-negative bacterium genome DNA and extraction method
A Gram-negative bacteria and extraction method technology, applied in the field of molecular biology, can solve the problems of large differences in nucleic acid extraction efficiency, achieve the effect of improving nucleic acid extraction efficiency, simple steps, and improving nucleic acid extraction purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
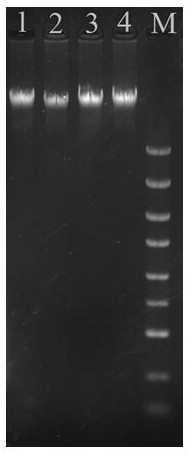
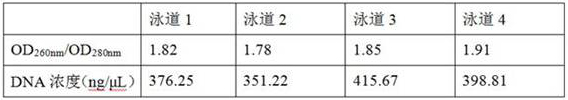
[0036] Example 1, a kit for rapidly extracting the genomic DNA of gram-negative bacteria, including a variety of working solutions, the specific preparation of each working solution is as follows:
[0037] (1) Working solution A: CTAB with a mass concentration of 0.8%, 2.5mol / L NaCl, a mass concentration of 2% Triton-X-100, 80mmol / L Tris-HCl (pH7.0-8.0), 15mmol / L EDTA , the mass concentration is 1% PVP, and the solvent is sterile deionized water.
[0038] (2) Working solution B: 2.0mol / L NaCl, 4.0mol / L guanidine isothiocyanate, 0.45mg / mL ribonuclease A, 0.07mol / L sodium citrate, the solvent is sterile deionized water.
[0039] (3) Working solution C is an ethanol solution with a volume fraction of 70%.
[0040] (4) Working solution D: 15mmol / L Tris-HCl, pH 7.0-8.0; solvent is sterile deionized water, pH 8.0.
[0041] (5) Magnetic bead binding solution: 1.5mol / L NaCl, 1.2mol / L guanidine hydrochloride, absolute ethanol with a volume ratio of 20%, isopropanol with a volume ratio ...
Embodiment 2
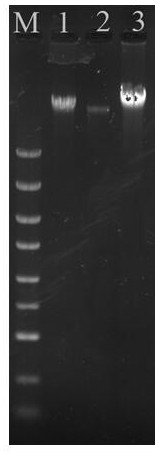
[0042] Embodiment 2, a kit for rapidly extracting the genomic DNA of Gram-negative bacteria, including a variety of working solutions, the specific preparation of each working solution is as follows:
[0043] (1) Working solution A: CTAB with a mass concentration of 0.5%, 3.5mol / L NaCl, Triton-X-100 with a mass concentration of 1%, 50mmol / L Tris-HCl (pH7.0-8.0), 25mmol / L EDTA, the mass concentration is 1% PVP, and the solvent is sterile deionized water.
[0044] (2) Working solution B: 1.5mol / L NaCl, 3.0mol / L guanidine isothiocyanate, 0.40mg / mL ribonuclease A, 0.1mol / L sodium citrate, the solvent is sterile deionized water.
[0045] (3) Working solution C is an ethanol solution with a volume fraction of 60%.
[0046] (4) Working solution D: 15mmol / L Tris-HCl, pH 7.0-8.0; solvent is sterile deionized water, pH 8.0.
[0047] (5) Magnetic bead binding solution: 0.5mol / L NaCl, 1.8mol / L guanidine hydrochloride, 30% absolute ethanol by volume, 60% isopropanol by volume, 3% polyeth...
Embodiment 3
[0048] Example 3, a kit for rapidly extracting the genomic DNA of Gram-negative bacteria, including a variety of working solutions, the specific preparation of each working solution is as follows:
[0049] (1) Working solution A: CTAB with a mass concentration of 1.0%, 1.5mol / L NaCl, Triton-X-100 with a mass concentration of 1%, 120mmol / L Tris-HCl (pH7.0-8.0), 8mmol / L EDTA, the mass concentration is 0.5% PVP, and the solvent is sterile deionized water.
[0050] (2) Working solution B: 1.0mol / L NaCl, 4.5mol / L guanidine isothiocyanate, 0.3mg / mL ribonuclease A, 0.08mol / L sodium citrate, the solvent is sterile deionized water.
[0051] (3) Working solution C is an ethanol solution with a volume fraction of 75%.
[0052] (4) Working solution D: 10mmol / L Tris-HCl, the solvent is sterile deionized water, pH8.0.
[0053] (5) Magnetic bead binding solution: 1.0mol / L NaCl, 1.5mol / L guanidine hydrochloride, 20% absolute ethanol by volume, 45% isopropanol by volume, 10% polyethylene gly...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| quality score | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com