A kind of preparation method of restriction endonuclease fsei
A technology of restriction endonuclease and encoding gene, which is applied in the field of preparation of restriction endonuclease FseI, can solve the problems of cumbersome separation and purification procedures, high protein price, low protein yield, etc. Market prospects, the effect of improving expression levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
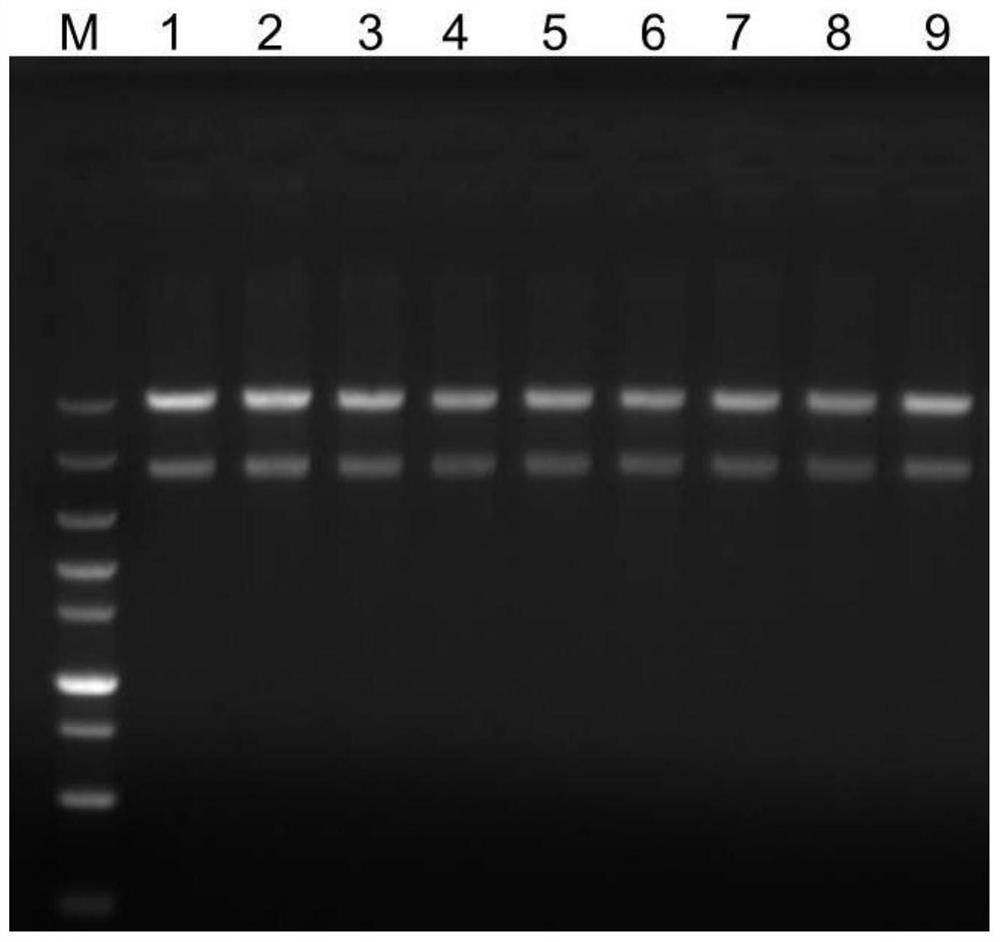
[0065] Example 1 Construction of pQE-80L-FseI prokaryotic expression plasmid
[0066] The codon-optimized FseⅠ gene was cloned into the pQE-80L vector containing 6×His (histidine) tag to construct the pQE-80L-FseⅠ prokaryotic expression plasmid. The specific method is as follows:
[0067] 1. Synthesis of FseⅠ gene
[0068] In order to realize the high-efficiency expression of FseI, the present invention firstly performs codon optimization on the coding gene of FseI according to the codon preference of Escherichia coli. In the process of codon optimization, the present invention found that the expression level and purity of FseI protein obtained by using conventional codon optimization software or simply following the codon preference of the E. coli expression system for codon optimization of FseI cannot satisfy the The enzyme was prepared in large quantities, and there was no obvious rule between the codon optimization position and optimization degree and the expression level...
Embodiment 2
[0091] Example 2 Expression of FseI restriction endonuclease
[0092] 1. Preparation of Escherichia coli Origami Competent Cells
[0093] (1) Pick the pre-stored Origami strain and streak it on empty LB solid medium, and cultivate overnight at 37°C;
[0094] (2) Pick a single clone colony in 25mL of empty medium, cultivate at 37°C for more than 12h, to OD 600 >1.5;
[0095] (3) Take 3mL of bacterial solution in 300mL empty LB medium (1:100) in the ultra-clean bench, shake it to OD at 18°C (200rpm) 600 is about 0.55;
[0096] (4) Bacterial liquid in ice bath for 10min, while pre-cooling Inoue transformation buffer;
[0097] (5) 4 °C, 4000rpm centrifugation for 10min to collect bacteria, discard the supernatant, invert on absorbent paper for 2min, blot dry the residual liquid, and resuspend the bacteria with 80mL of Inoue transformation buffer;
[0098] (6) 4 °C, 4000rpm centrifugation for 10min to collect bacteria, discard the supernatant, invert on absorbent paper for 2...
Embodiment 3
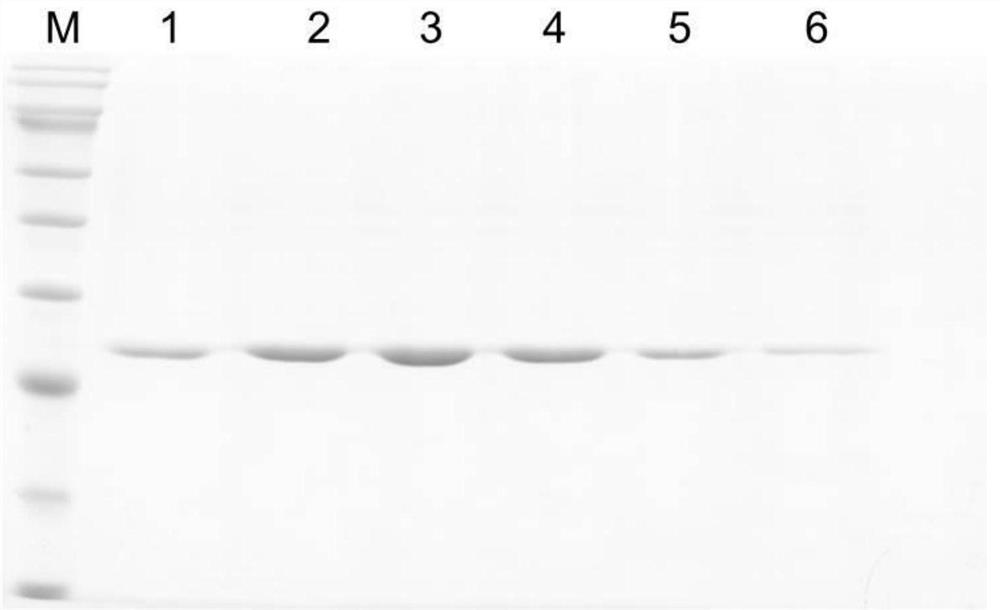
[0120]Example 3 Purification of FseI restriction endonuclease
[0121] 1. Protein purification by nickel column
[0122] (1) Use a 0.22 μm filter to filter the supernatant;
[0123] (2) Incubate the supernatant with Beads at 4°C for 1-2 hours, at which time FseI protein binds to Beads;
[0124] (3) adding the supernatant into the purification column, allowing the liquid to flow out naturally under the action of gravity, and the target protein bound to the Beads remains in the purification column, and the purification column is always kept in the chromatography cabinet to maintain a low temperature environment;
[0125] (4) Wash the purification column with Wash Buffer (20mM Tris-HCl, pH8.2; 25mM imidazole; 150mM NaCl) for 5-10 column volumes to remove the impurity proteins that are not bound to Beads, and use fast staining reagents to detect proteins in real time until When the quick-staining reagent no longer turns blue, it can be judged that no impurity protein has been el...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com