Primer and probe for detecting ZNF384-related fusion gene by multiple fluorescence PCR technology, and application
A technology of EP300-ZNF384 and ARID1B-ZNF384, which is applied in the fields of life science and biology, can solve problems such as insufficient sensitivity, high cost of FISH reagents, and easy contamination of the process, and achieve the effects of less sample demand, improved efficiency, and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
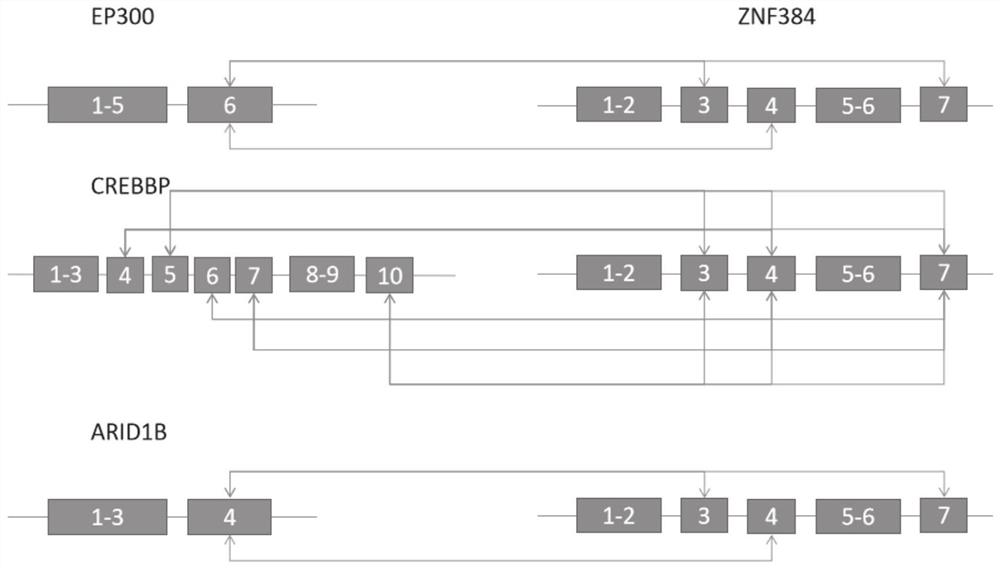
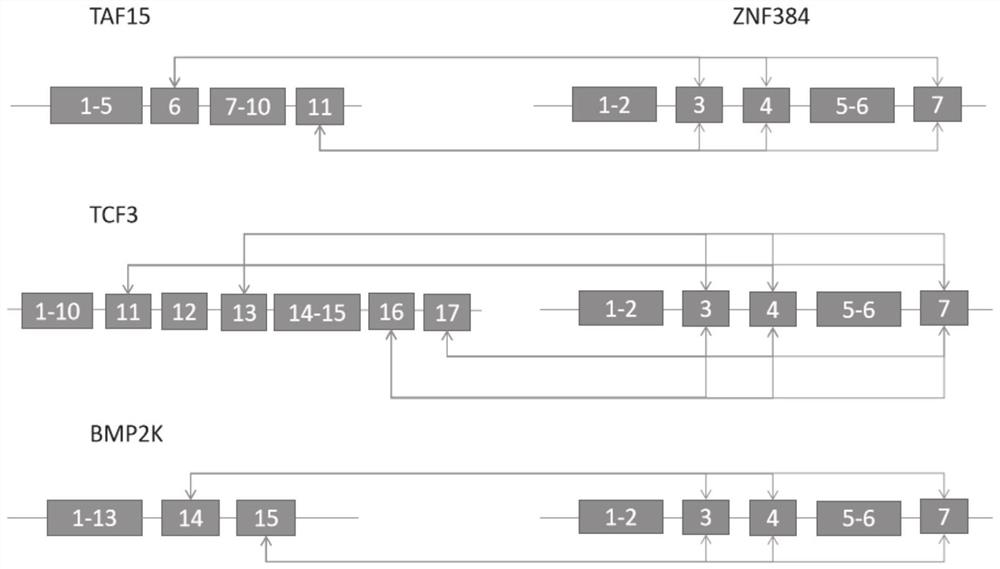
[0047] Embodiment 1: The amplification primer probes of ZNF384-related fusion genes of the present invention are shown in the following table:
[0048]
Embodiment 2
[0050] Extraction of blood RNA:
[0051] Add 0.5ml of whole blood sample to 1ml of erythrocyte lysate, mix by inverting, leave at room temperature for 5min, then centrifuge at 4000rpm for 3min; discard the supernatant; resuspend in erythrocyte lysate, repeat washing twice to observe the volume of cell clumps, reaching 10-30ul. Add 1 ml Trizol Reagent and pipette repeatedly until there are no cell clumps in the lysate, and let stand at room temperature for 5 minutes. Add 200 μl of chloroform to the above Trizol solution, vortex and mix until fully emulsified, let stand on ice for 10 minutes, and then centrifuge at 14,000 rpm for 10 minutes. Take 450 μl of the supernatant and transfer it to a centrifuge tube filled with pre-cooled isopropanol, mix up and down, put it on ice at -30°C for 10 minutes, then centrifuge at 14,000 rpm for 10 minutes, discard the supernatant, add 1 ml of 75% ethanol to wash the precipitate, and then 14,000 rpm Centrifuge for 5 min; discard the supernat...
Embodiment 3
[0054] Multiplex fluorescent PCR amplification:
[0055] Configure the reverse transcription system according to the following reagents and reagent volume, Primer mix 1ul, 5*RT buffer 4ul, Enzyme Mix 1ul (Toyobo (Shanghai) Biotechnology Co., Ltd.); add deionized water 10ul; finally add sample RNA template 4ul. The reverse transcription program is: 37°C for 60 minutes, 98°C for 5 minutes. Sample cDNA was prepared.
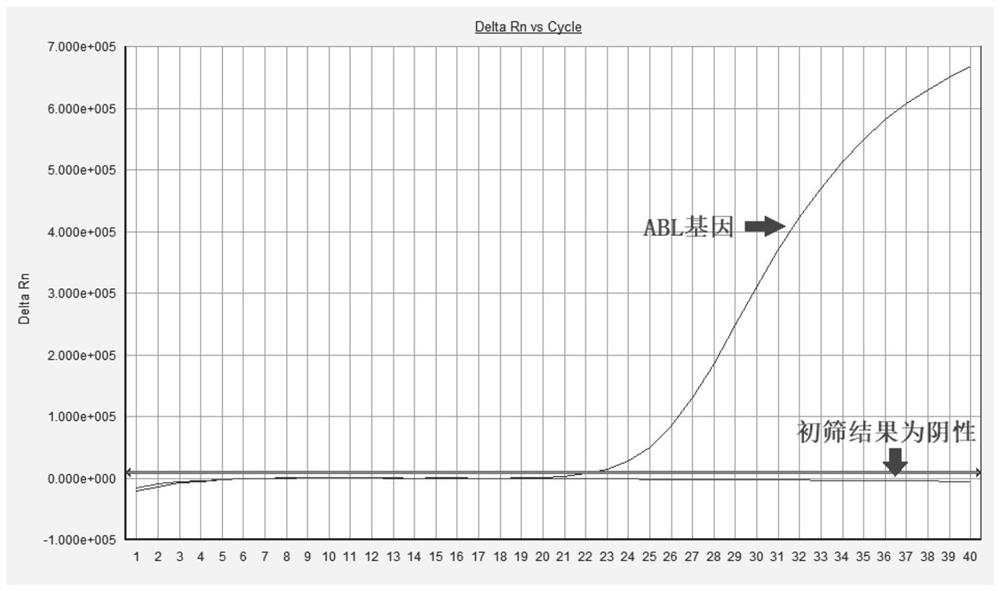
[0056] Configure the PCR amplification system according to the following reagents and reagent volumes, each primer of the primary screening system is 0.1ul, and each probe is 0.05ul; THUNDERBIRD Probe qPCR Mix 12.5ul, 50*ROX 0.5ul (Toyobo (Shanghai) Biotechnology Co., Ltd.); Add 8 ul of deionized water; finally add 2 ul of cDNA template; the amplification program is as follows: pre-denaturation at 95°C for 1 min; denaturation at 95°C for 10 sec, extension at 58°C for 35 sec, 40 cycles, and fluorescence signal collection set at 58°C for extension.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com