Prokaryote-derived argonaute protein and application thereof
A prokaryotic and protein technology, applied in the field of molecular biology, can solve problems such as short synthesis cycle, and achieve the effects of short synthesis cycle, cost saving and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
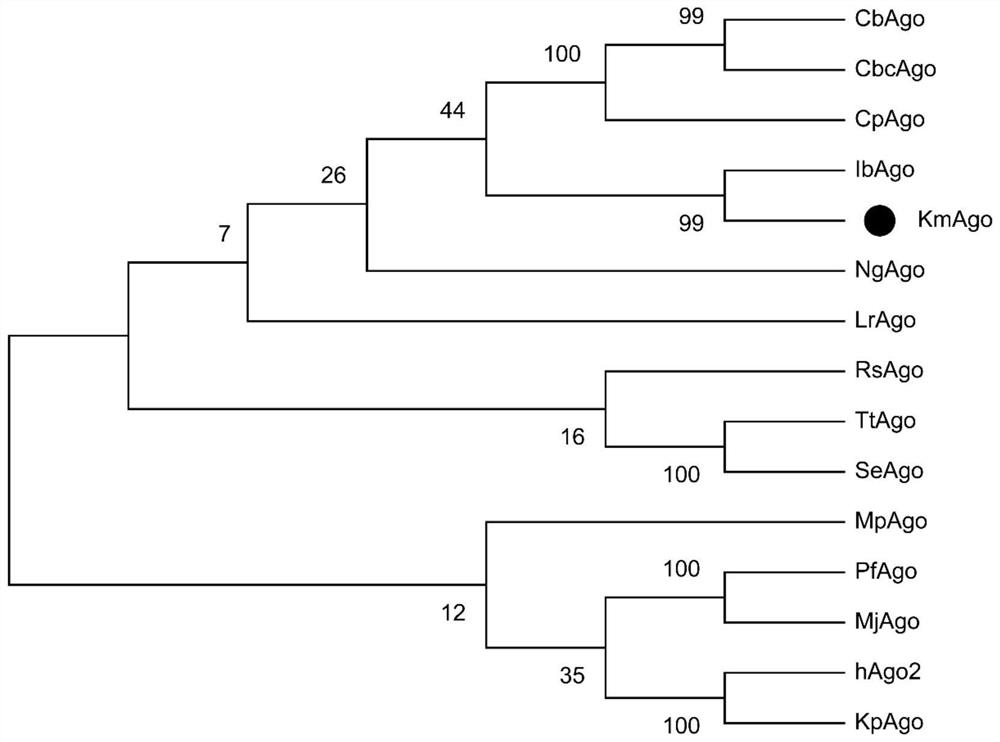
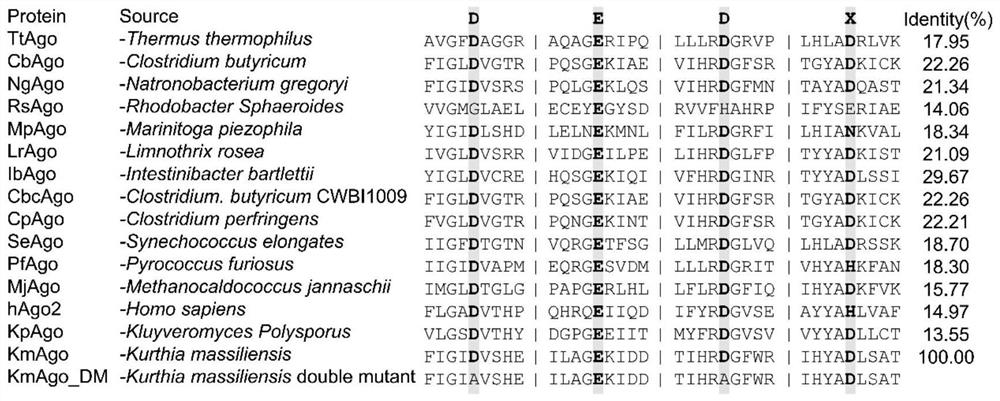
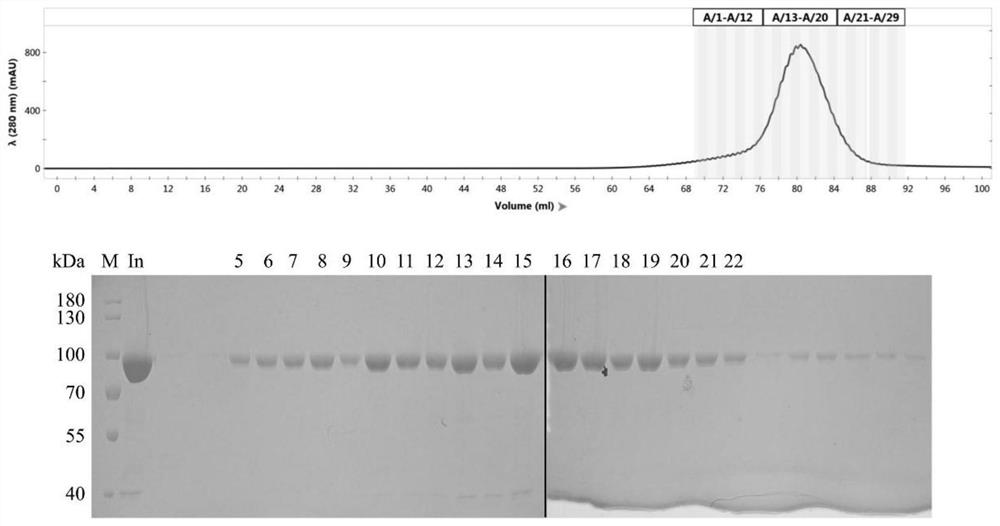
[0107] Example 1 KmAgo expression and purification
[0108] Transform the pET28a-KmAgo plasmid into Escherichia coli BL21(DE3), inoculate a single colony into LB liquid medium containing 50 mg / mL kanamycin, and culture it on a shaker at 37°C and 220 rpm. When the OD600 of the bacteria reaches At 0.8, move to a shaker at 18°C overnight. Collect the cells by centrifugation at 6000 rpm for 10 min, wash with Buffer A (20 mM Tris–HCl pH 7.5, 500 mM NaCl, 10 mM imidazole), resuspend the cells in Buffer A, add PMSF at a final concentration of 1 mM, and press broken. Centrifuge at 18000 rpm for 30 min and collect the supernatant. After the supernatant was filtered, Ni-NTA purification was performed.
[0109] Wash 9 column volumes with 10 mM imidazole and 20 mM imidazole (add in 3 times), wash 3 column volumes with 50 mM, 80 mM, 100 mM, 150 mM, 200 mM, 250 mM, 300 mM, 1 M, and take samples Carry out SDS-PAGE detection. Collect the eluted fraction containing the high-purity targe...
Embodiment 2
[0111] Example 2 Testing the cleavage activity of KmAgo
[0112] To assess which combinations of RNA / DNA guides and RNA / DNA targets KmAgo was able to cleave, activity assays were performed for all possible combinations. Cleavage assays were all performed at 37 °C at a 4:2:1 (pAgo:guide:target) molar ratio. Place 800 nM KmAgo with 400 nM guide in a solution containing 10 mM HEPES-NaOH pH 7.5, 100 mM NaCl, 5 mM MnCl 2 and 5% glycerol in reaction buffer and incubated at 37°C for 10 min for guide loading. Nucleic acid targets were added to 20OnM. After 1 h reaction at 37°C, the reaction was terminated by mixing the sample with 2x RNA loading dye (95% formamide, 18 mM EDTA and 0.025% SDS and 0.025% bromophenol blue) and heating at 95°C for 5 min. The lysate was resolved by 20% denaturing PAGE, stained with SYBR Gold (Invitrogen) and stained with Gel Doc TM XR+ (Bio-Rad) visualization.
[0113] The product band (34 nt) was not observed in DNA / RNA (guide / target) control assays ...
Embodiment 3
[0115] Example 3 KmAgo can be used at extremely low Mn 2+ cleavage of target RNA
[0116] Subsequent activity assays focused on finding the Mn for which KmAgo exhibits guide-directed target RNA cleavage within 15 min 2+ concentration range. mn 2+ The concentration range is set to 0-10 mM, and when the guide is 5’ phosphorylated DNA, the target RNA can be cut efficiently when the Mn2+ concentration is 0.01 mM ( Figure 7 ); when the guide is 5’ phosphorylated RNA, when the concentration of Mn2+ is greater than 0.5 mM, the target RNA can be effectively cut ( Figure 8 ).
[0117] Figure 7 DNA guides were incubated with KmAgo for 10 minutes at room temperature before adding the target. A 34 nt product band appeared between 0.05-10 mM, indicating an efficient reaction.
[0118] Figure 8 RNA guides were incubated with KmAgo for 10 min at room temperature prior to addition of target. A 34 nt product band appeared between 0.05-10 mM, indicating an efficient reaction.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com