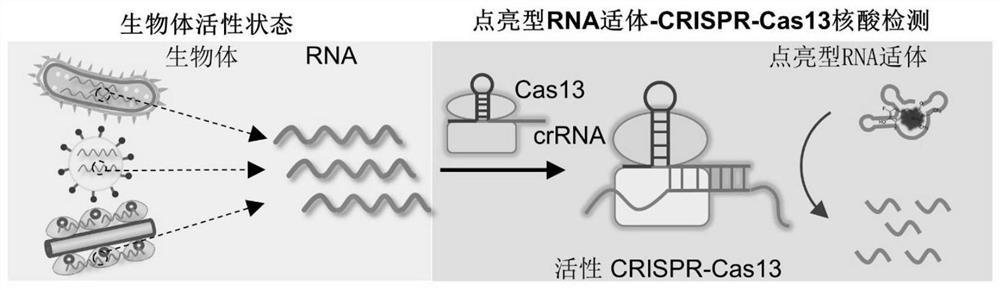
CRISPR-Cas13 nucleic acid detection kit based on lightened RNA aptamer
A detection kit and the technology of the kit are applied in the field of nucleic acid detection, which can solve the problems of complicated operation process and high detection cost, and achieve the effects of simplified operation process, high sensitivity and cost saving.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1 Preparation of light-up RNA aptamer and signal recognition probe crRNA
[0069] In this example, the Broccoli aptamer was used as an example to prepare a bright-type RNA aptamer, and to prepare CRISPR-RNA (crRNA). The light-up RNA aptamer Broccoli aptamer can specifically bind to the nucleic acid dye DFHBI-1T as a beacon for Cas13 digestion. The signal recognition probe crRNA can specifically bind to the target RNA and play the role of molecular recognition.
[0070] Use the method of in vitro transcription to obtain Broccoli aptamer and crRNA, the steps are as follows:
[0071] Into a 100 μL centrifuge tube, add 10 μL 10×phi29 DNA polymerase buffer, 10 μL L-Broccoli aptamer, 10 μL promoter, denature at 90°C for 3-10 minutes, and react at room temperature for 30-90 minutes. Add 3 μL phi29 DNA polymerase, 2 μL dNTP mixture, and extend it to complete complementarity at 30°C for 30-90 minutes. Finally, add 20 μL of 5× transcription buffer, 2 μL of T7 RNA polym...
Embodiment 2
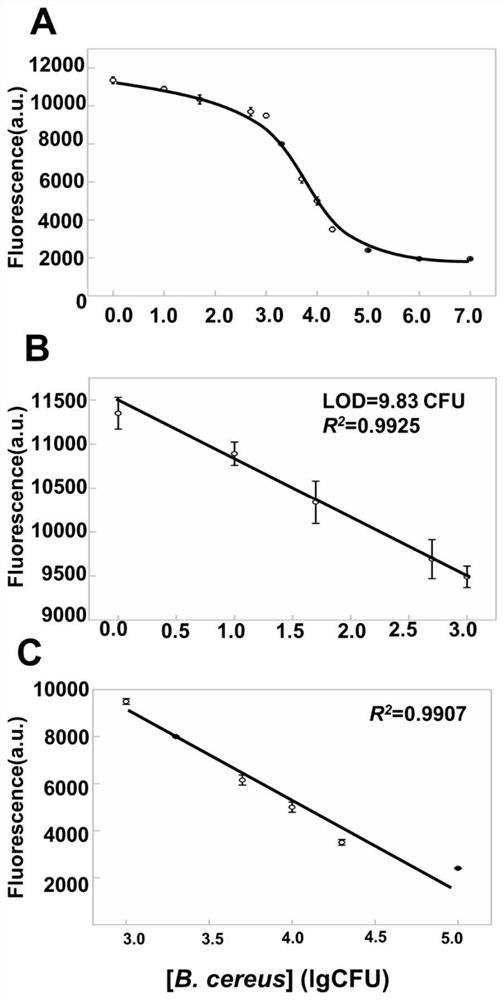
[0076] Embodiment 2 draws standard curve
[0077] Proceed as follows:
[0078]① Cultivate Bacillus cereus (B. cereus) at 37°C until the late logarithmic growth phase, and prepare live bacterial solutions with different concentrations using the gradient coefficient of normal saline to extract RNA.
[0079] ② Take 10 μL of the Broccoli aptamer prepared in Example 1, 3 μL of crRNA, and 4 μL of DFHBI-1T, add the RNA solution extracted from different concentrations of Bacillus cereus obtained in ①, and add 0.3 μL of Cas13 solution (1 pmol / μL) , add 19.7μL of water, and react at 37°C for 30min.
[0080] ③ Fluorescence was detected at an excitation wavelength of 468nm and an emission wavelength range of 498-560nm, with a detection step of 1nm, and the fluorescence value at the concentration of viable bacteria was recorded.
[0081] ④ Use the concentration of Bacillus cereus as the abscissa and the fluorescence value at this concentration as the ordinate to draw a standard curve. T...
Embodiment 3
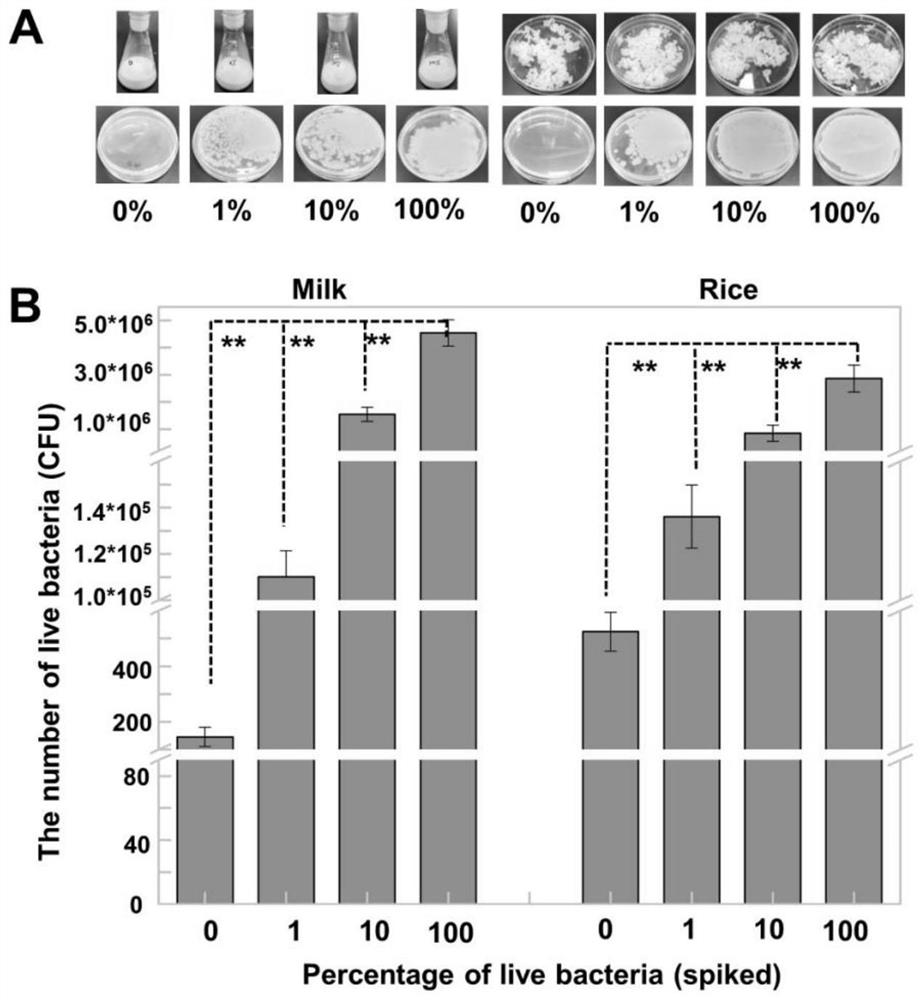
[0083] Bacillus cereus quantity detection in embodiment 3 milk and rice
[0084] Add Bacillus cereus with different concentrations of viable bacteria to milk and rice, detect the number of viable bacteria in the sample after 48 hours of culture, and analyze the significant difference between each group and the sample with 100% viable bacteria. The steps are as follows:
[0085] ① Take 40g of cooked rice and 40ml of commercially available pure milk and put them into 4 Erlenmeyer flasks, and sterilize at 121°C for 20min.
[0086] ② Dilute the Bacillus cereus in the late logarithmic growth period to 10% with 0.85% normal saline. 5 CFU / mL.
[0087] ③ Centrifuge the bacterial solution prepared in ②, resuspend the bacterial solution with 70% isopropanol and normal saline respectively, treat at room temperature for 1 hour, centrifuge, and resuspend with normal saline to obtain dead bacteria and live bacteria.
[0088] ④ Take the viable and dead bacteria liquids prepared in ③, and m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com