Bacteria identification and typing analysis genome database and identification and typing analysis method
A bacterial identification and genome technology, applied in the field of bacterial genome identification and typing, can solve the problems of identification result deviation, identification threat, genome draft integrity and contamination are not easy to distinguish, and achieve the effect of increasing speed and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
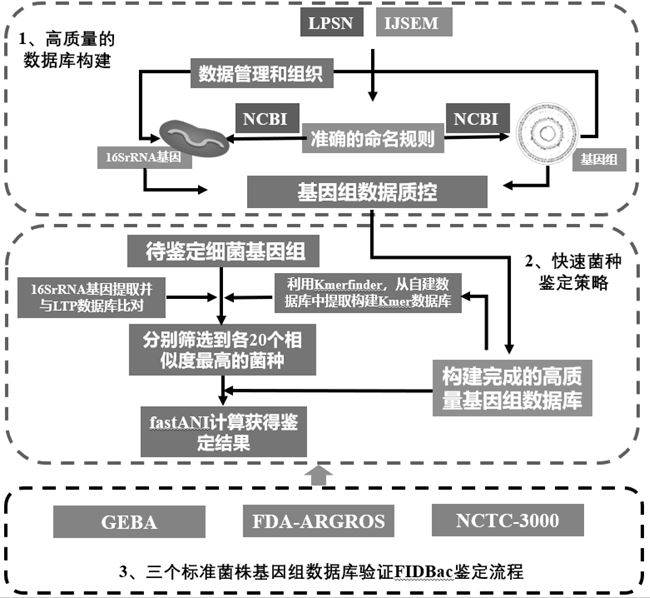
[0035]As shown in FIG. 1, the bacterial identification and classification of the present invention analyzes the genomic database MDBACDB construction process as follows:
[0036]1) Collect bacterial genome information from NCBI, and collect the correspondence forms of "strain", "cultured", "cloned", and "annotation", establish genomic information and meta information, and clarify the source of each genome.
[0037]2) Get a list of effective published bacterial names and type strains from LPSN, and review Bergey's ate and bacterial system manuals and iJSem's articles. After the screening, the qualified strain bacteria genome entered the database for management.
[0038]3) After screening the bacterial genomic sequence of the warehouse, the MDBACDB construction is completed. Filter the wrong, low-quality bacterial genome by self-contained Python program MDBacqctools.
[0039]The steps of MDBACQCTools are under quality control: First, the integrity and pollution rate of each genome are evaluated u...
Embodiment 2
[0043]As shown in FIG. 1, the FiDBAC analysis of the bacterial genomic data analysis platform of the present invention is as follows:
[0044]1) Get a genomic sequence of public bacteria GCF_008121515.1_genomic.fna;
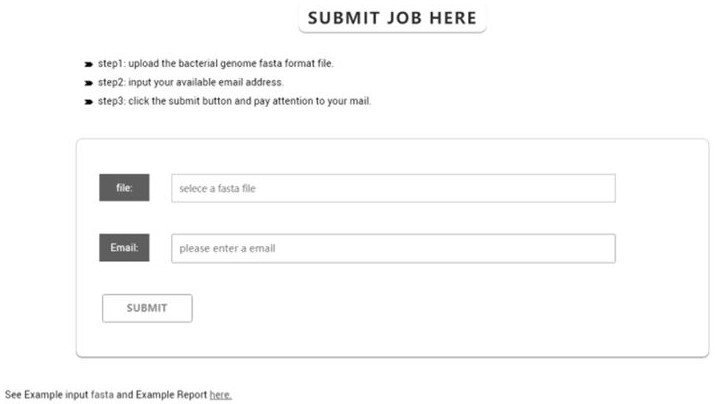
[0045]2) Submit to bacterial genomic data analysis platform FIDBAC (image 3), Identification analysis by self-compiled Python program FIDBAC.
[0046]FiDBAC's analysis process is as follows: First, the 16S rRNA sequence to be identified to be identified (GCF_008121515.1_genomic.fna) is compared with the LTP database; secondly, use Kmerfinder (V3.1), from GCF_008121515.1_genomic.fna The K-MERS is compared with the MDBACDB's K-MER database; again, obtain the top 20 bacterial ID numbers obtained in the first two steps, and extract genomic sequences from the bacterial genome database MDBACDB, using FASTANI (V1. 1) Calculate the ANI value of the query genome and the corresponding type of strain genome; Finally, the identification result returns only the closest species and the ANI v...
Embodiment 3
[0048]As shown in FIG. 1, the FiDBAC analysis of the bacterial genomic data analysis platform of the present invention is as follows:
[0049]1) Get Staphylococcus Capitis from NCBI,Bacillus CEREUS,Bacillus AnthracisGenome sequence GCA_001650475.1, GCA_002564865.1 and GCA_000725325.1;
[0050]2) Extract the 16S rRNA gene sequence from the genome, using a 16S rRNA gene sequence and the reference database LTP to perform BLAST comparison identification, and sort it according to the score value.
[0051]3) Submit to the bacterial genome data analysis platform FIDBAC (image 3), Identification analysis by self-compiled Python program FIDBAC. The Fidbac analysis process is as follows: First, the 16S rRNA sequences in the bacterial genome (GCA_001650475.1.FNA, GCA_001650475.1.FNA, GCA_002564865.1.FNA) are extracted to compare the LTP database; secondly, using Kmerfinder (V3 .1), comparison from the K-MERS extracted from GCF_008121515.1_Genomic.fna with MDBACDB's K-MER database; again, obtain the top...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com