Method for measuring tissue distribution of MSC
A technology of tissue distribution and real-time fluorescence quantification, which is applied in the field of biomedical detection, can solve the problem of inability to accurately quantify the number of mesenchymal stem cells in tissues, and achieve the effects of low detection limit, accurate quantification and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Determination of detection primers
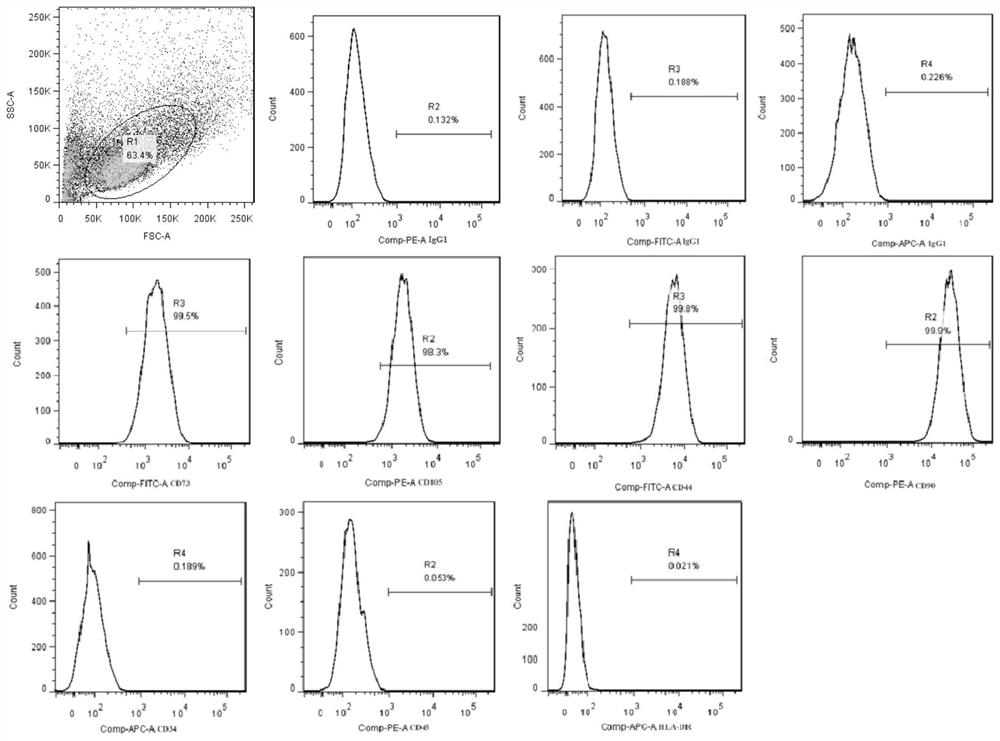
[0024] (1) Will figure 1 1×10 human umbilical cord-derived mesenchymal stem cells with positive rates of CD73, CD105, CD44, and CD90 detected by flow cytometry above 98%, and negative rates of CD34, CD45, and HLA-DR below 0.2% 7 For each, use blood / cell / tissue genomic DNA extraction kit (DP304, Tiangen Biochemical Technology (Beijing) Co., Ltd.) to extract DNA, calculate the total amount of DNA, and then half-dilute 9 times to obtain 10 serial concentrations of DNA standards solution;
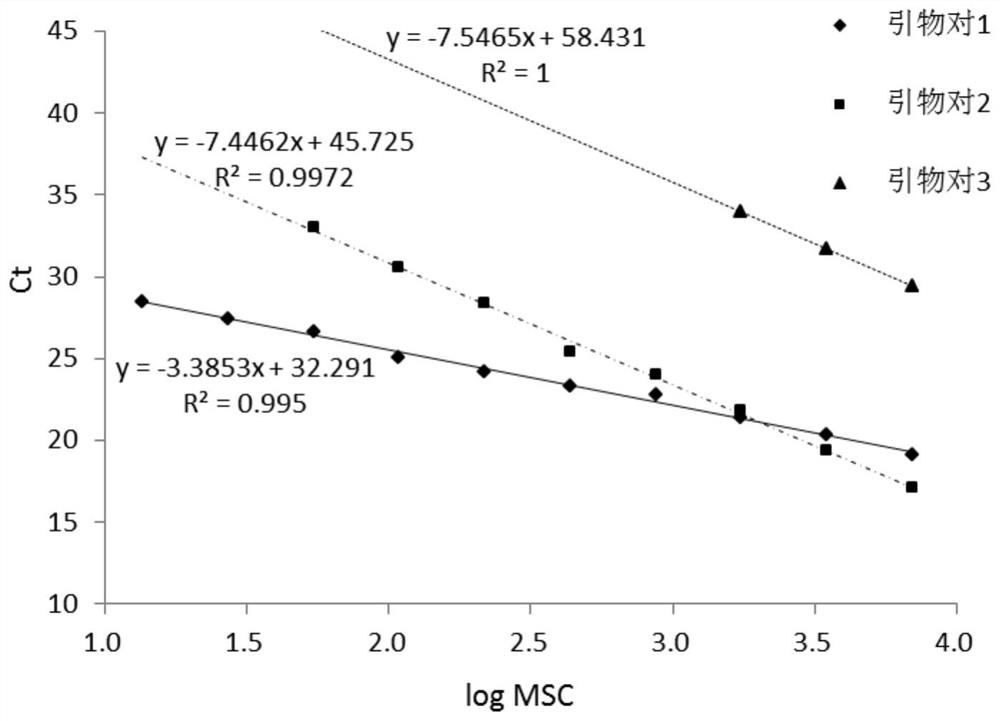
[0025] (2) According to the sequence of SEQ ID NO:7-9, design three pairs of primers as shown in the table below and synthesize them:
[0026]
[0027] Use the DNA standard solution in step (1) as a template to perform real-time fluorescent quantitative PCR, and the reaction system is as follows:
[0028]
[0029] The amplification reaction is as follows:
[0030]
[0031] The melting curve acquisition reaction is as follows: 95...
Embodiment 2
[0034] Example 2 Repeatability detection
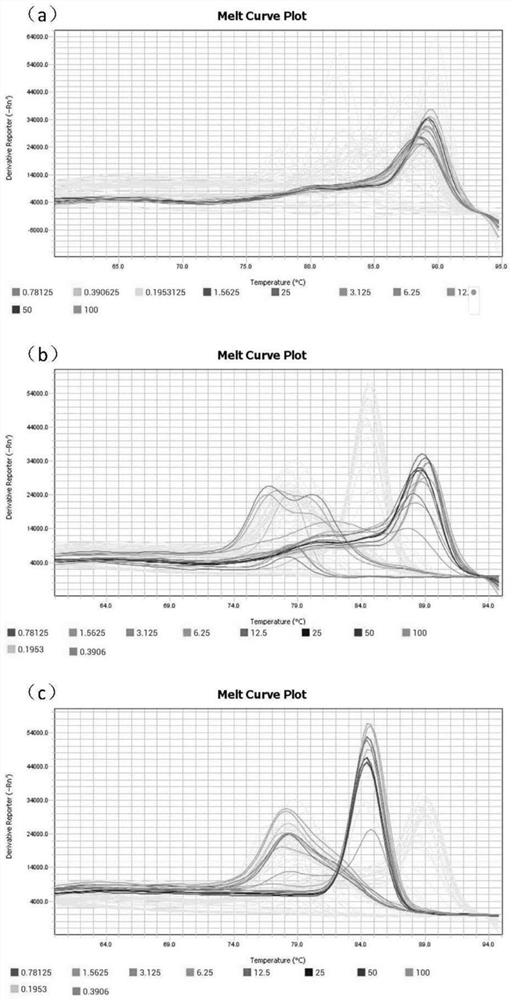
[0035] The first pair of primers was tested for repeatability, and the DNA standard solutions of 10 serial concentrations in Example 1 were also taken, and the amplification experiment was carried out according to the method in Example 1, and the melting curve obtained was as follows: Figure 4 As shown, it can be seen from the melting curve that primer pair 1 has no non-specific amplification, which can be used to evaluate the method, and the coefficient of variation (CV) is calculated according to the Ct value of each standard solution. When the amount of DNA changes from 0.19ng to 100ng, the coefficient of variation is from 0.97% to 2.76%, which is less than 3%, which can be used for subsequent quantitative detection.
Embodiment 3
[0036] Example 3 Quantitative detection of tissue distribution of mesenchymal stem cells
[0037] (1) Will figure 1 The positive rates of CD73, CD105, CD44, and CD90 detected by flow cytometry are above 98%, and the negative rates of CD34, CD45, and HLA-DR are below 0.2%. The cell density in the liquid is 1×10 7 cells / mL, BALB / c-nude nude mice (female mice) were used as test subjects, injected into the tail vein, each injection volume: 200μL, 15 mice in total;
[0038] At 24h, 48h, 72h, 96h, and 120h after injection, 3 nude mice were randomly selected at each time point and killed by necking after anesthesia. The heart, liver, spleen, lung, and kidney tissues were collected, washed with blood, sucked water, and weighed. Then freeze and store at -80°C;
[0039] After all the samples were taken, DNA was extracted using the Blood / Cell / Tissue Genomic DNA Extraction Kit (DP304, Tiangen Biochemical Technology (Beijing) Co., Ltd.);
[0040] (2) Using the sequence shown in SEQ ID ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com