Method for constructing microscale sample m6A modification detection library under assistance of Tn5 transposase and application thereof
A technology of transposase and sequencing library, which is applied in the field of RNA modification site detection, can solve the problems of inability to meet the needs of trace samples, limitations, and limited applications, and achieves the goal of reducing input, reducing costs, and avoiding contamination of sequencing data. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0068] Example 1 A Tn5 transposase-assisted high-throughput sequencing method for detecting m6A modifications in trace samples
[0069] The primers (Vazyme, cat.no.TD202) needed in this example are as follows (N5-index includes N501-508, N7-index includes N701-712):
[0070]
[0071]
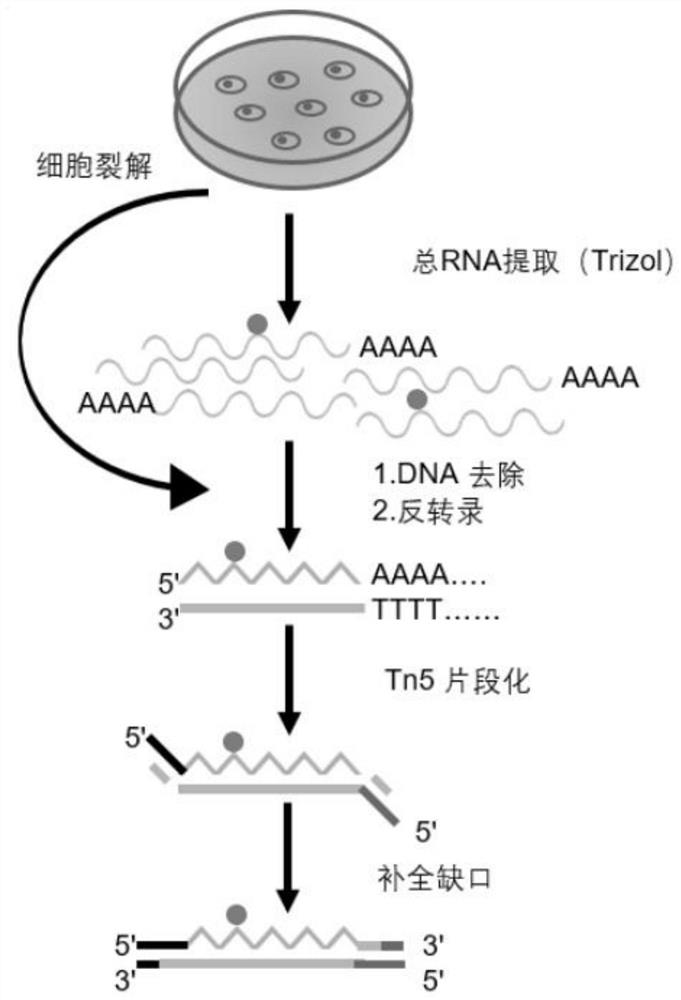
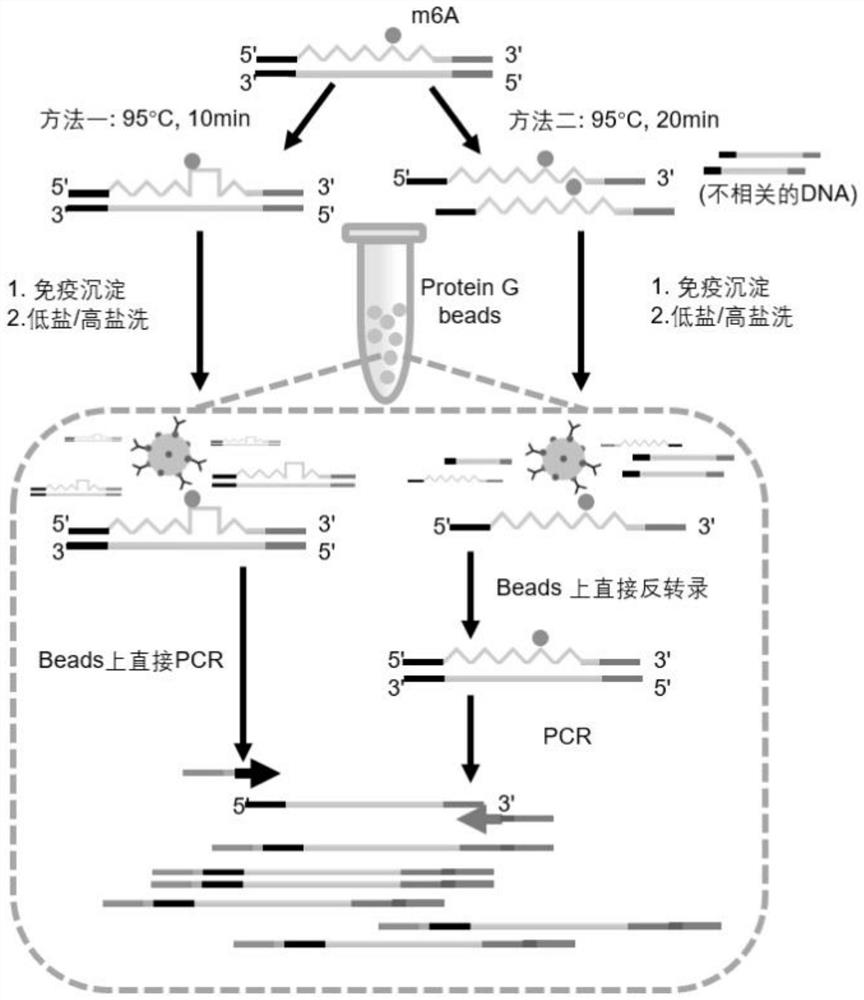
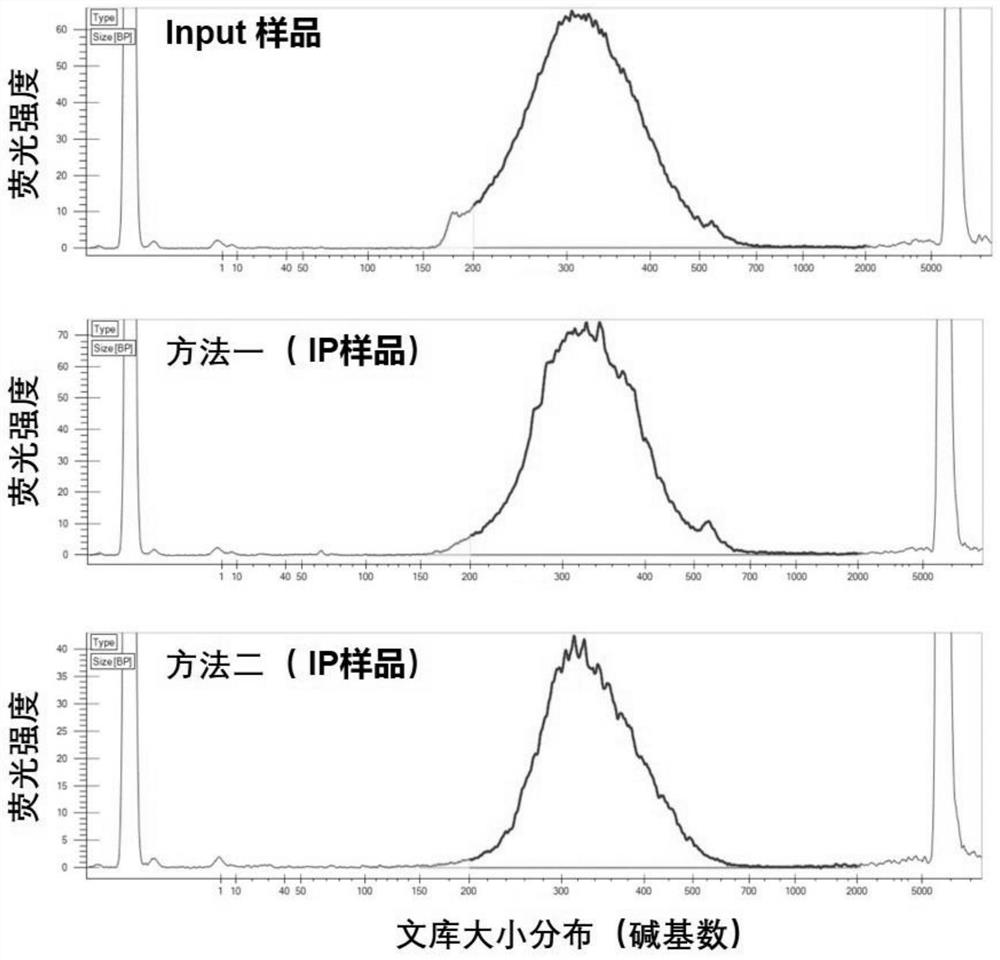
[0072] The total RNA required in this embodiment is 0.06 micrograms, and the process of steps (1)-(7) is as follows figure 1 Shown, the flow process of steps (8)-(16) is as follows figure 2 Shown:
[0073] (1) Using a carbon dioxide incubator, place 293T cells (purchased from the Cell Bank of the Chinese Academy of Sciences) in modified DMEM medium (DMEM; 273 Corning, cat.no.10-017-CV), and add 10% (v / v ) fetal bovine serum (FBS; Gibco), 5% CO 2 , and cultured at 37°C until the number of cells reached the level of 100,000.
[0074] (2) Discard the cell culture medium, wash twice with PBS, digest the cells with trypsin, centrifuge the cell suspension (1000rpm / 5min), discard the supern...
Embodiment 2
[0123] Example 2 A Tn5 transposase-assisted high-throughput sequencing method for detecting m6A modification in trace samples
[0124] The detection sample in this embodiment is 293T cells (2000 cells), that is, the cell lysate is used as the detection sample.
[0125] The primers used in this example are the same as those in Example 1.
[0126] The process of steps (1)-(7) is as follows figure 1 Shown, the flow process of steps (8)-(16) is as follows figure 2 Shown:
[0127] (1) Using a carbon dioxide incubator, 293T cells were placed in modified DMEM medium (DMEM; 273 Corning, cat. no. 10-017-CV) and 10% (v / v) fetal bovine serum (FBS; Gibco) was added.
[0128] (2) Discard the cell culture medium, wash twice with PBS, digest the cells with trypsin, centrifuge the cell suspension (1000rpm / 5min), discard the supernatant, reselect the cells with PBS, and then use flow cytometry Sorter, sort out 2000 cells.
[0129] (3) Add 3 μL of cell lysate to the cells sorted in step ...
Embodiment 3
[0134] Example 3 A Tn5 transposase-assisted high-throughput sequencing method for detecting m6A modification in trace samples
[0135] The detection sample in this embodiment is a drop of blood sample (5 microliters).
[0136] The primers used in this example are the same as those in Example 1.
[0137] The operation flow of this embodiment is as follows Figure 6 Shown:
[0138] (1) Obtain 5 microliters / sample of fingertip peripheral blood from healthy volunteers using a blood collection needle.
[0139] (2) Using erythrocyte lysate (Roche, 11814389001) to remove erythrocytes in peripheral blood to obtain a large number of leukocytes.
[0140] (3) Total RNA was extracted using the classic Trizol (Thermo Fisher Scientific, catalog no. 15596026) method, and operated according to the instructions to obtain total RNA.
[0141] (4) The following m6A modification detection method is the same as that in Example 1, wherein step (11) adopts method two.
[0142] The m6APeak identi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com