Method for mining microorganism anti-heavy metal function based on conserved sequence clustering analysis
A conservative sequence and anti-heavy metal technology, applied in the field of microorganisms, can solve problems such as high failure probability, heavy workload, and high degree of genetic variation, and achieve accurate mining results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
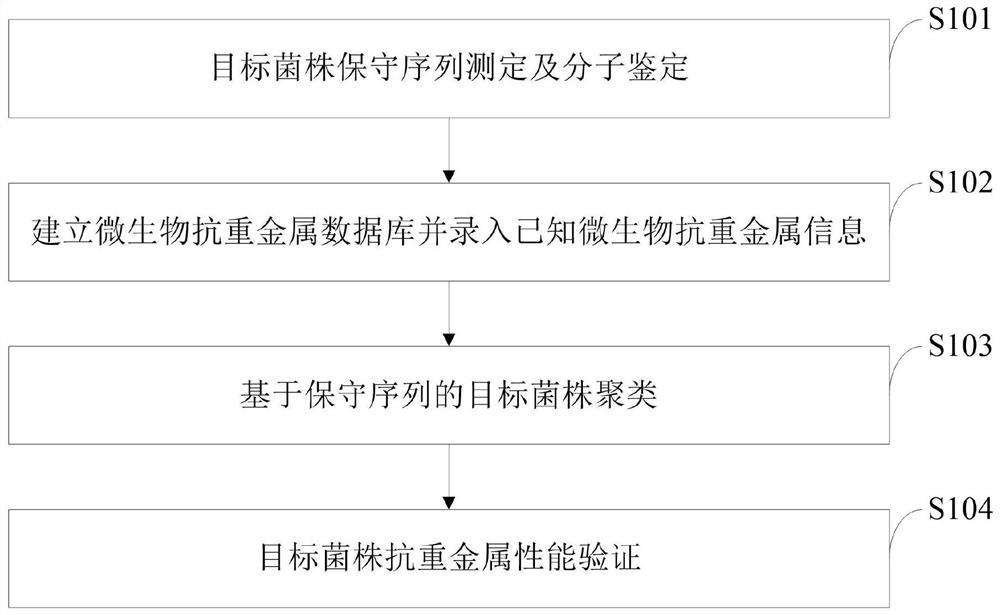
[0048] The implementation process of the microbial anti-heavy metal function mining method based on conservative sequence cluster analysis provided by the embodiments of the present invention is as follows:
[0049] (1) Conserved sequence determination and molecular identification of the target strain. Different microorganisms have different conserved sequences at different taxonomic levels. In this step, recognized conserved sequences, such as bacterial 16S sequence, fungal ITS sequence, mitochondrial COX1 gene sequence, etc., can be used, or the target strain can be determined by comparing the known sequence information of the family or genus to which the target strain belongs. Specific conserved sequence segments. After determining the conserved sequence segment, determine the conserved sequence of the target strain. Establish a phylogenetic tree to re-determine the species of the target strain.
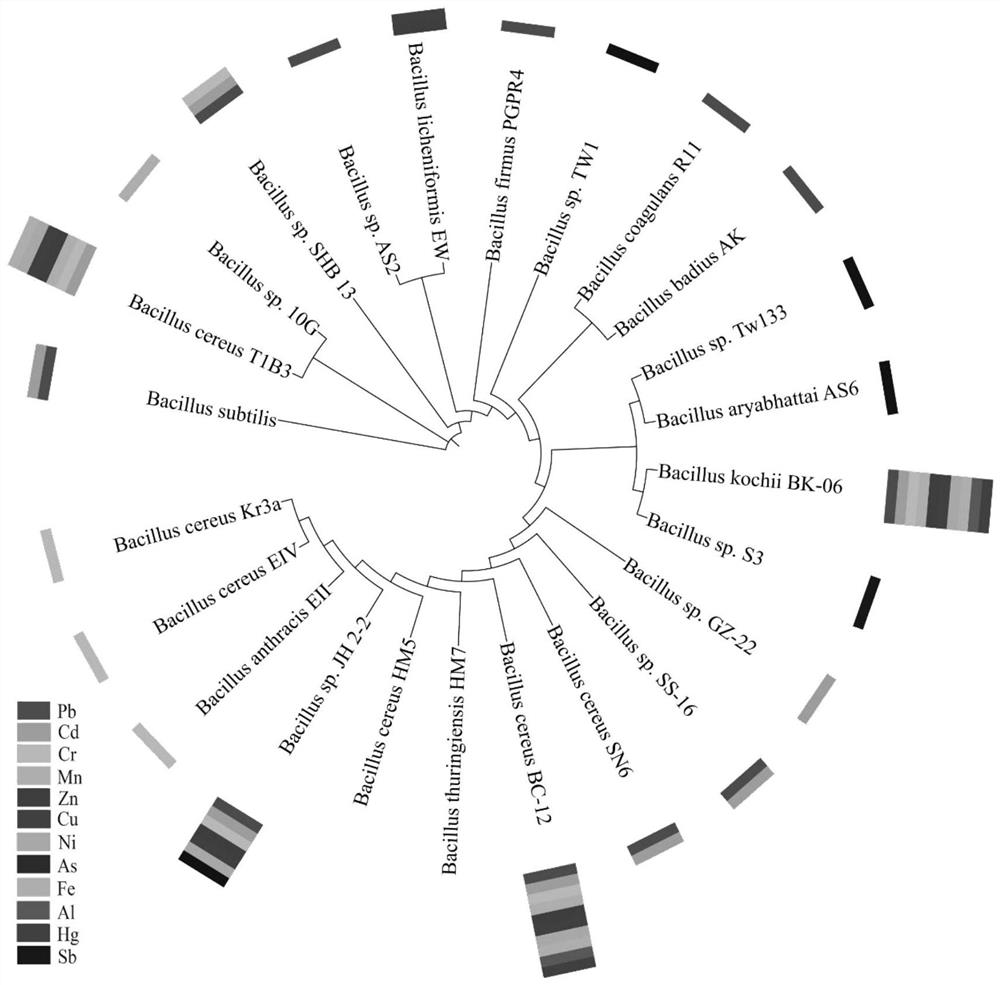
[0050] (2) Establish a microbial anti-heavy metal database and enter known...
Embodiment 2
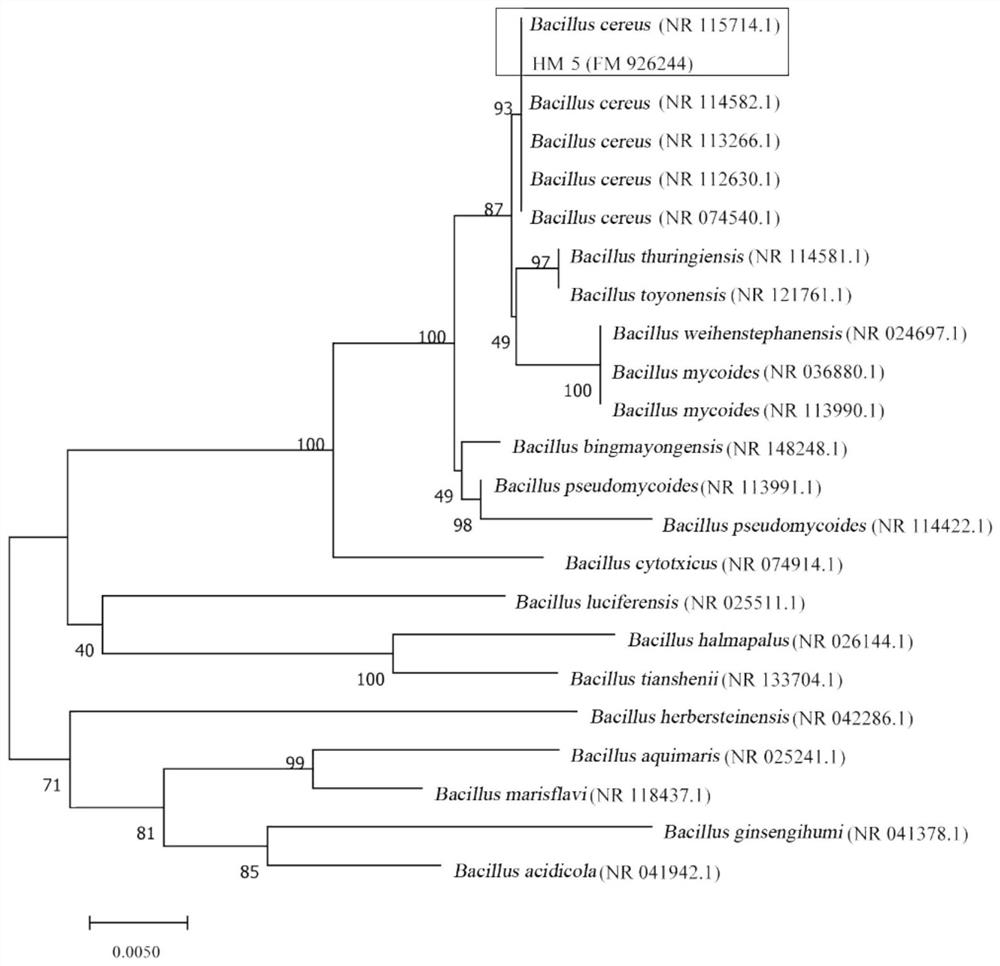
[0054] (1) Conserved sequence determination and molecular identification. Select a bacterium of unknown species and unknown function, select the 16S rDNA, which is recognized as a conserved sequence by the bacterium, as the conserved sequence, use the kit to extract the DNA of the bacterial strain, use the extracted total DNA as a template, and use 16S rRNA primers for PCR amplification. The primer is: 27FAGAGTTTGATCCTGGCTCAG, and the reverse primer is: 1492RGGTTACCTT GTTACGACTT. 20ul Taq reaction system, including 10×TaqBuffer 2ul, 2mM dNTPs 2ul, 25mM MgSO4 1.2ul, Taq enzyme 1ul, Primer1 (10pm) 1ul, Primer2 (10pm) 1ul, Plate 1ul, PCR enhancer 5ul, H2O 5.8ul, after amplification sequencing. The 16S sequence of the bacterium was determined, and the determination result is shown in sequence 1. Species phylogenetic relationship was identified based on conserved sequences, and species phylogenetic trees were constructed. The bacterium was identified as Bacillus cereus ( figur...
Embodiment 3
[0063](1) Conserved sequence determination and molecular identification. Two strains of fungi with similar morphology were obtained from the slag of an antimony mine. The genomic DNA of the fungus was extracted, and the ITS region of the tolerant fungus was amplified by PCR using the forward primer ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and the reverse primer ITS4 (5'-TCCTCCGTTATTGATATGC-3'). The PCR amplification system conditions are (50 μL): 2×TaqPCR mix 25 μL (including Taq DNA polymerase, dNTPs, MgCl2, reaction buffer), Forward primer (10 μmol L-1) 2 μL, Reverse primer (10 μmol L-1) 2 μL, Template 1μL, ddH2O 20μL. PCR cycling conditions: 98°C 2min; 98°C 10s; 54°C 10s, 72°C 10s, 35 cycles; 72°C 5min. After the PCR amplification products were sequenced, the ITS sequences of the above-mentioned fungi were determined, and the sequence information is shown in Table 2. The obtained gene sequence was subjected to BLAST analysis in National Center for Biotechnology Information (NCBI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com