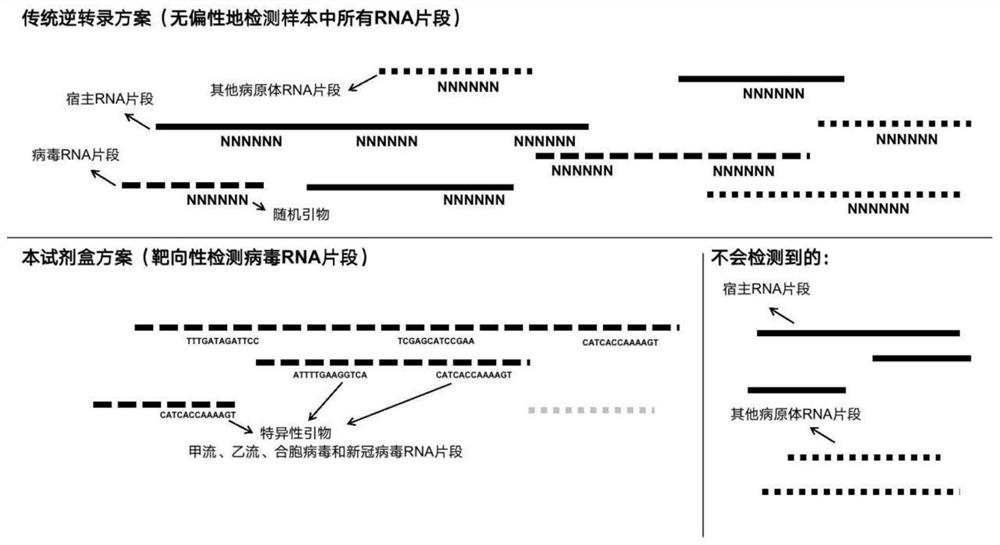
A kind of ngs primer set and kit for detecting respiratory virus pathogens
A technology of primer sets and kits, which is applied in the fields of life sciences and biology, can solve the problems of reduced ability to detect viral pathogens and complex nucleic acid sources, and achieve the effects of improving detection sensitivity, avoiding base preference, and simple and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1: detect the specific reverse transcription primer design of flow A, flow B, syncytial virus and novel coronavirus (SARS-CoV-2)
[0043] First, download the reference genome sequences of influenza A, influenza B, syncytial virus and new coronavirus from the NCBI GeneBank nt database.
[0044]After multiple sequence alignment of these genome sequences, use PYFASTA (http: / / pypi.python.org / pypi / pyfasta / ) to cut each genome into 400-base long fragments, allowing 200 bases between these fragments base overlap. Then search for kmer (13mer) of 13 bases in the window region of 50 bases at the beginning and end of these 400-base long fragments (it can be the sense strand or the antisense strand), and all kmers are based on the appearance The frequency is sorted in descending order, and the 13mer with the highest frequency (the most conservative sequence) is selected as the candidate primer selected in the first round, and then all 500-base long fragments containing ...
Embodiment 2
[0047] Using the sensitivity reference products in the national reference products for detection of influenza A, influenza B, syncytial virus and new coronavirus nucleic acid of the National Institute of Inspection and Quarantine, the effect of using 594 virus-targeting primers and using random primers for metagenomic library sequencing was tested.
[0048] 1) Source of samples: Standard Plate of Respiratory Pathogens of China Institute of Inspection and Quarantine, and New Coronavirus Standard.
[0049] 2) Sample pretreatment method: In order to simulate the alveolar lavage fluid sample, a certain amount of human cells need to be mixed into the pure virus culture to provide the host nucleic acid background. Use 1 × 10 for the fixed value of A flow, B flow, syncytial virus and SARS-CoV-2 sensitivity reference 5 Immortalized human leukocytes / mL were diluted to 100,000 copies / mL and marked as S1; then the cells of this concentration were used to make a 2-fold concentration gradi...
Embodiment 3
[0109] NGS testing was performed on 4 real sputum / throat swab samples with nucleic acids of influenza A, influenza B, syncytial virus, and new coronavirus to verify the effect of the reverse transcription primer set.
[0110] 1) Sample source and nucleic acid extraction: sputum / throat swab samples from 4 patients with the target virus, passed through Qiagen Total RNA extracted with Viral RNA extraction kit.
[0111] 2) The experimental steps of reverse transcription and library construction are the same as in Example 2.
[0112] 3) All libraries were single-ended 75bp sequencing (SE75) using the illumina Nextseq platform, the host sequence was removed by bioinformatics analysis, and the number of reads detected by the new coronavirus and the genome comparison rate after splicing with parameters were analyzed, as follows surface:
[0113]
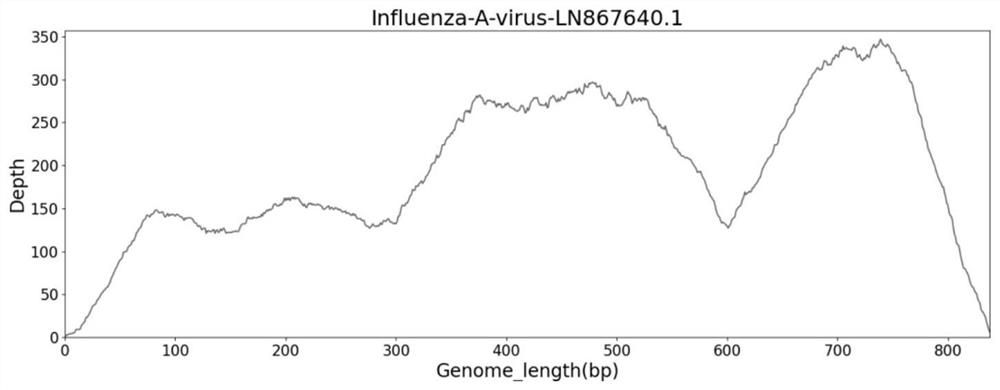
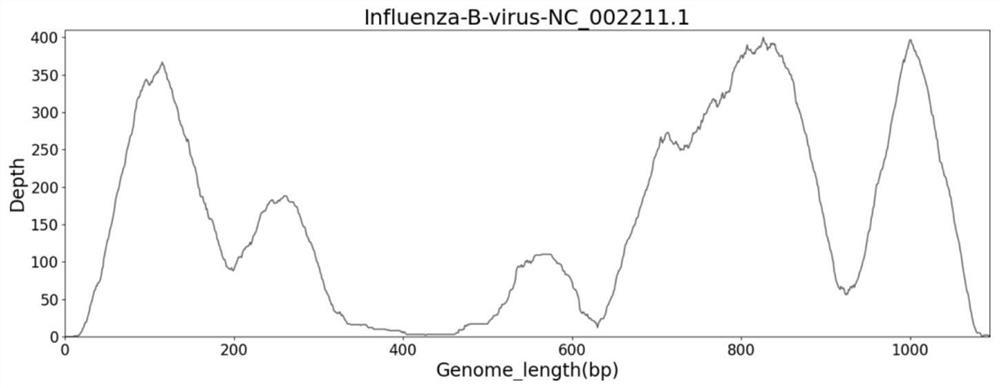
[0114] figure 2 is the genome coverage map of influenza A in the sample, image 3 is the B genome coverage map in the sample, Fi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com