Dynamic monitoring system for iPSC differentiation
A promoter, SV40 technology, applied in the field of cell biology, can solve problems such as difficult to track differentiation efficiency, difficult to large-scale culture, hindering iPSC application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
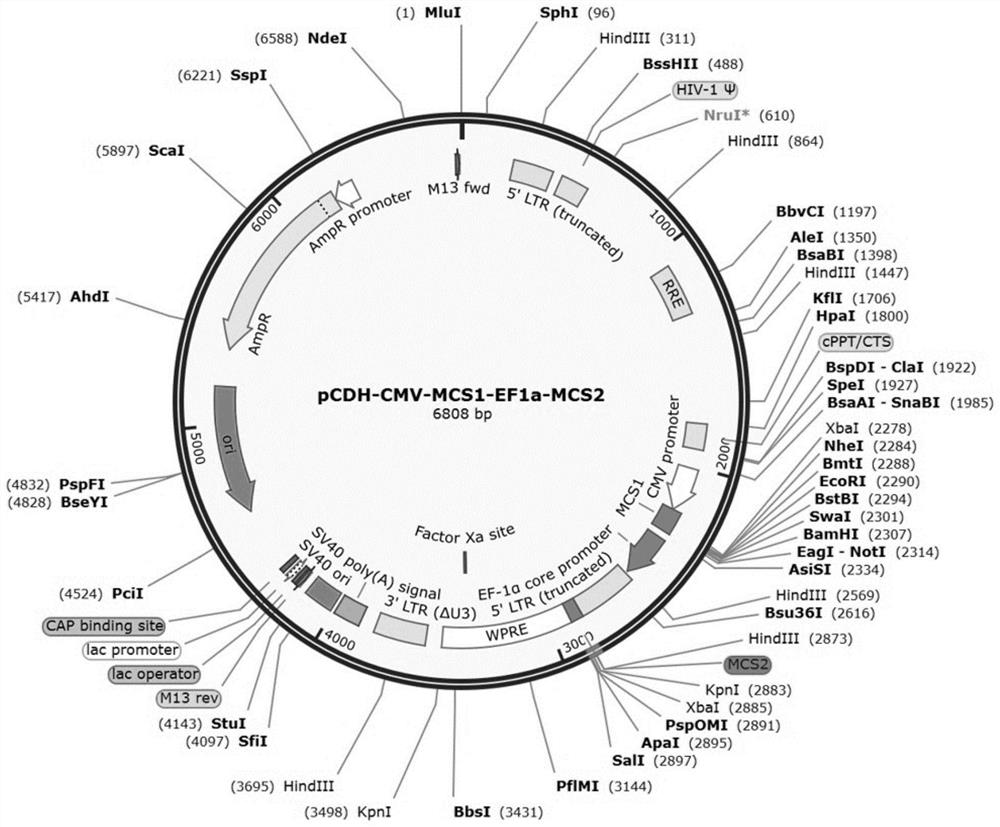
[0124] Example 1 pCDH-CMV-MCS1-EF1a-MCS2 plasmid construction
[0125] 1. Experimental materials
[0126] The plasmid vector pCDH-CMV-MCS-EF1a-Puro used in the examples of the present invention was purchased from Invitrogen.
[0127] 2. Construction of pCDH-CMV-MCS1-EF1a-MCS2 plasmid
[0128] The fragment including EF1-Puro was digested from the site on the pCDH-CMV-MCS-EF1a-Puro vector. Select NotI for the 5' end of the restriction site and SalI for the 3' end. Through gene synthesis, the EF1a-5'LTR (truncated)-MCS2 sequence was inserted into the above restriction plasmid, the EF1a sequence information is shown in SEQ ID NO: 1, and the 5'LTR (truncated) sequence information is shown in SEQ As shown in ID NO: 2, the sequence information of MCS2 is shown in SEQ ID NO: 3. MSC2 includes restriction enzyme cutting sites HindIII, KpnI, XbaI and ApaI in sequence.
[0129] 3. Experimental results
[0130] The experimental results show that the map of the constructed pCDH-CMV-MCS...
Embodiment 2
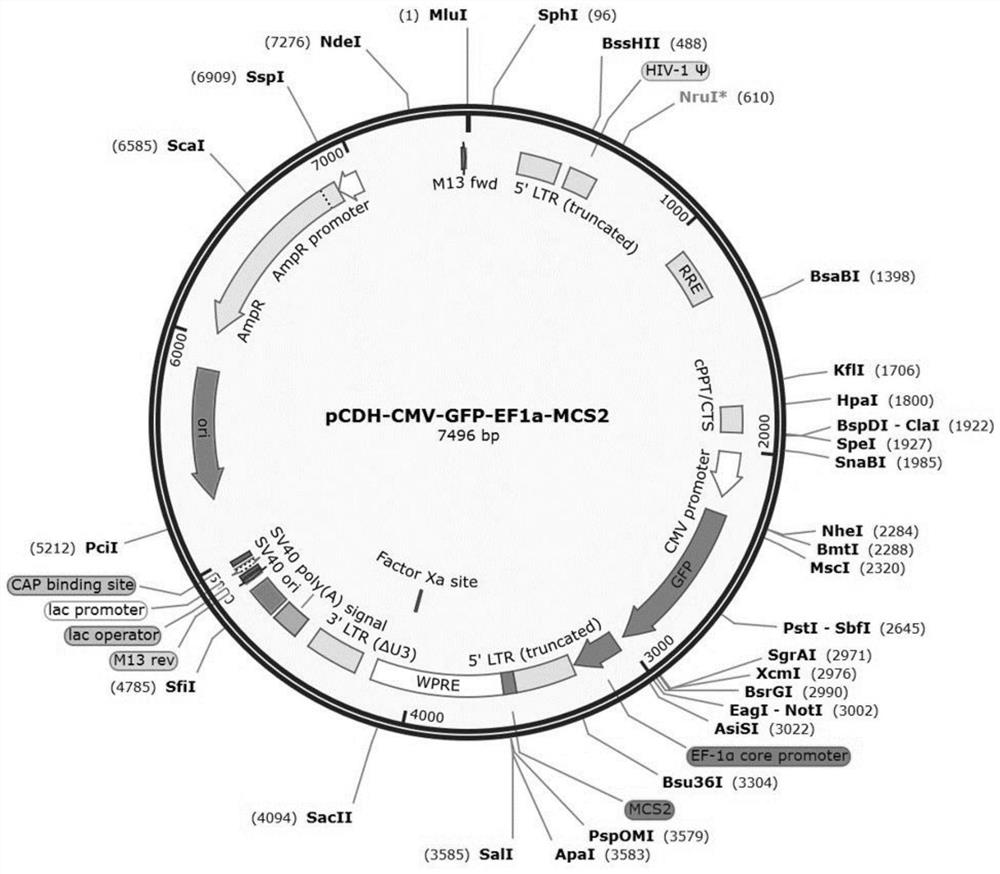
[0131] Embodiment 2 pCDH-CMV-GFP-EF1a-MCS2 plasmid construction
[0132] 1. Construction of pCDH-CMV-GFP-EF1a-MCS2 plasmid
[0133] Select restriction sites from the MCS1 sequence on the pCDH-CMV-MCS1-EF1a-MCS2 vector, and cut the vector into line segments. NheI (GCTAGC) was selected for the 5' end of the restriction site, and NotI (GCGGCCGC) was selected for the 3' end. Through gene synthesis, the GFP sequence was inserted into the restriction plasmid, wherein the GFP sequence information is shown in SEQ ID NO:4. GFP is green fluorescent protein, and the GFP sequence can be replaced with the sequence of other fluorescent proteins, such as: red fluorescent protein, yellow fluorescent protein, blue fluorescent protein, cyan fluorescent protein, orange fluorescent protein, etc., or these fluorescent protein derivatives Therefore, replacing the GFP sequence with the sequence of other fluorescent proteins or derivatives thereof is also included in the protection scope of the pre...
Embodiment 3
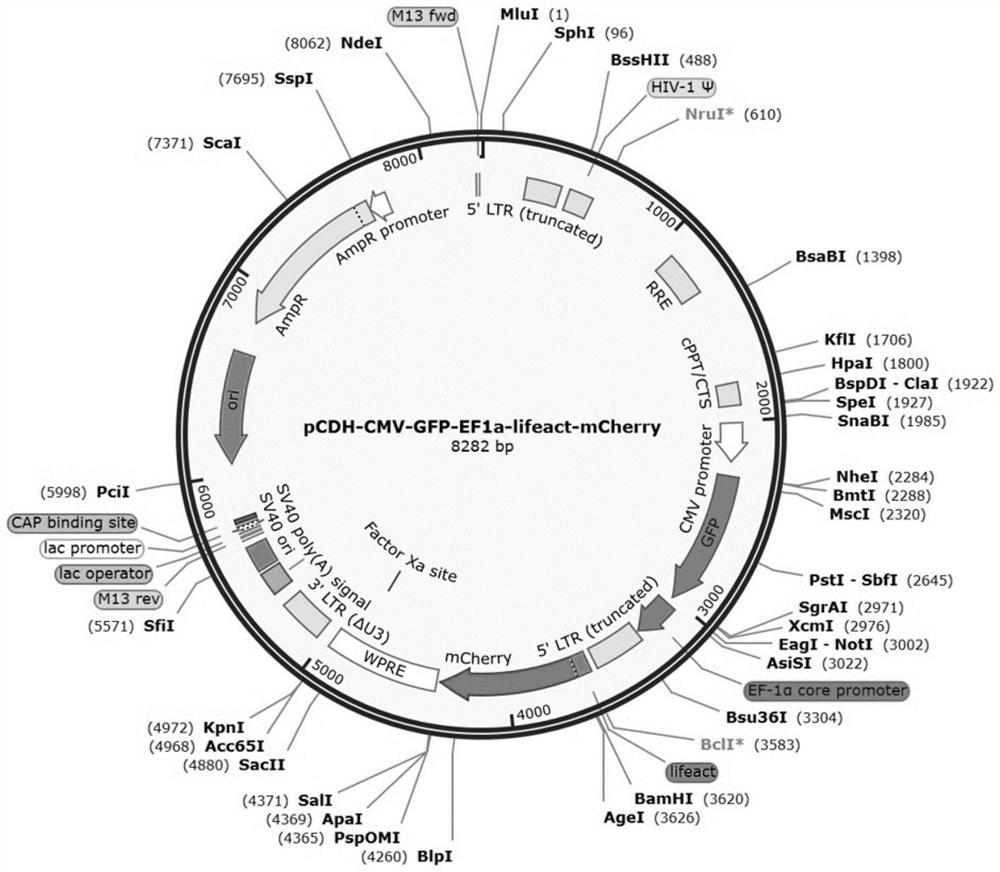
[0136] Example 3 pCDH-CMV-GFP-EF1a-lifeact-mCherry plasmid construction
[0137] 1. Construction of pCDH-CMV-GFP-EF1a-lifeact-mCherry plasmid
[0138] Select restriction sites from the MCS2 sequence on the pCDH-CMV-GFP-EF1a-MCS2 vector, and cut the vector into line segments. HindⅢ (AAGCTT) was selected for the 5' end of the restriction site, and XbaI (TCTAGA) was selected for the 3' end. Through gene synthesis, the lifeact-mCherry sequence is inserted into the above restriction plasmid, wherein the lifeact sequence information is shown in SEQ ID NO:5, and the mCherry sequence information is shown in SEQ ID NO:6. mCherry is a derivative protein of red fluorescent protein, and the mCherry sequence can be replaced by other fluorescent protein sequences, such as: red fluorescent protein, yellow fluorescent protein, blue fluorescent protein, cyan fluorescent protein, orange fluorescent protein, etc., or these fluorescent proteins The sequence of the derivative protein, in order t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com