Chitosan deacetylase combined mutant with optimal pH reduction and application
A technology of deacetylase and chitobiose, which is applied in the field of bioengineering, can solve the problems of poor stability of the product GlcN, reduce the optimal pH of enzyme catalysis, etc., and achieve the effect of improving conversion rate and enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Site-directed mutant library construction for predicted sites
[0044] The amino acid sequence of chitobiose deacetylase (as shown in SEQ ID NO.1) derived from extreme thermophilic archaea was subjected to site-directed mutation, the mutations were specifically G74D, H152E, F168A, W232A, and the gene encoding the mutant (sequence shown in SEQ ID NO.2) was connected with the p43NMK vector to obtain the recombinant expression vector p43NMK-M14.
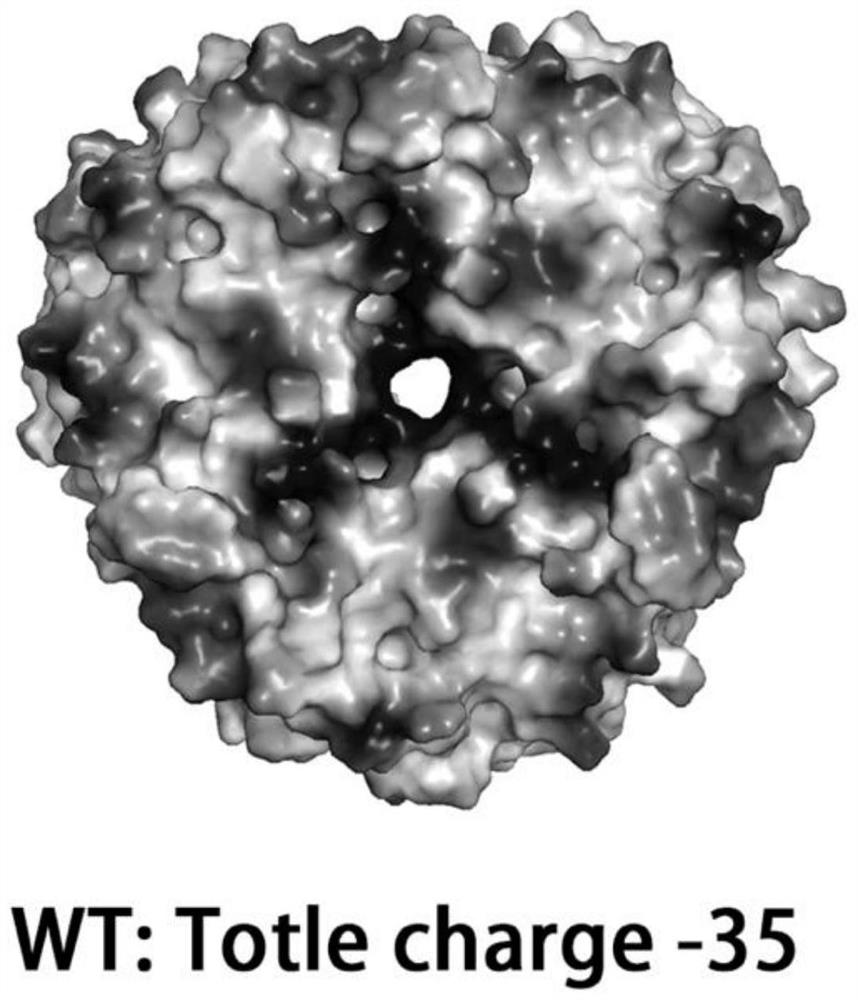
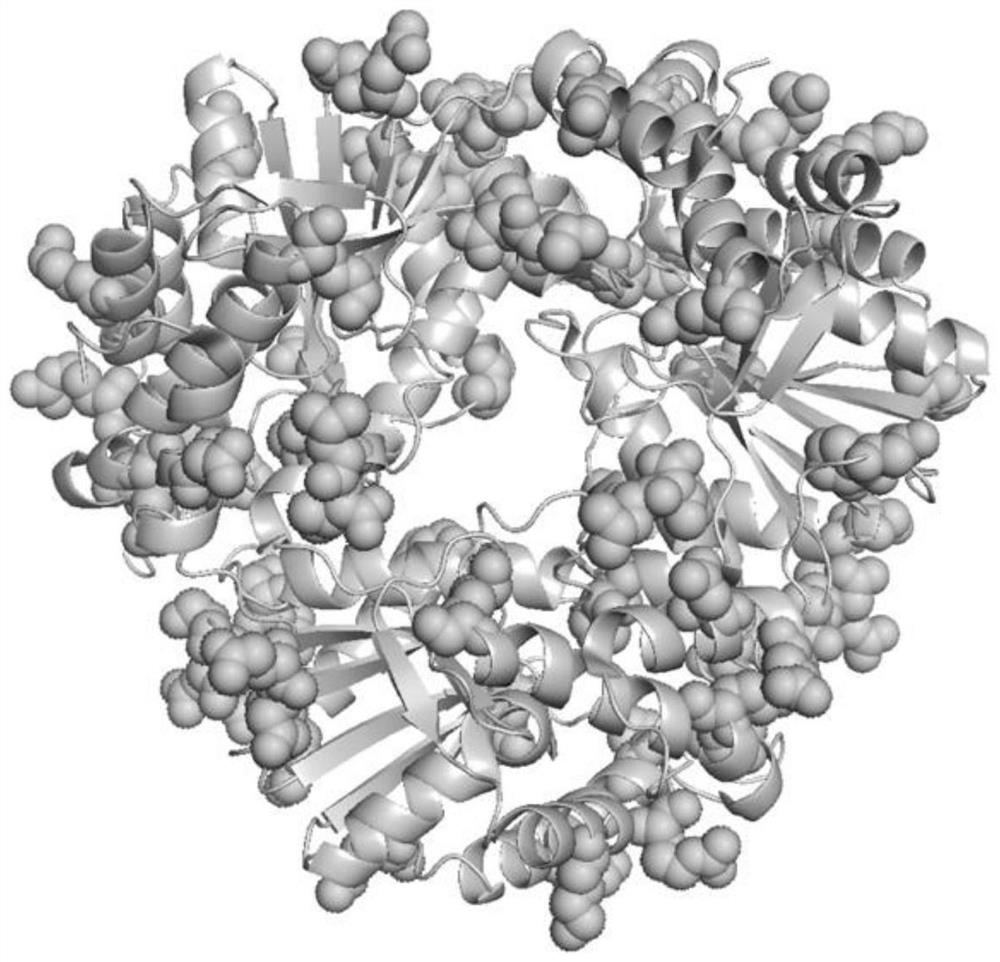
[0045] The surface charge of Dac wild type and the prediction of related sites are as follows figure 1 and 2 , the initial surface charge is -35, and the predicted 23 sites are Q29, N30, H86, K94, K106, N112, S121, K134, N176, T177, I181, N184, N187, S188, K215, R221, I228, K231, K246, H264, L271 and I272. Using p43NMK-M14 as a template, design site-directed mutagenesis primers, as shown in Table 1, for PCR amplification and Escherichia coli BL21 transformation.
[0046] Table 1 Primer Sequence
[0047]
[0048] ...
Embodiment 2
[0050] Embodiment 2 high-throughput screening
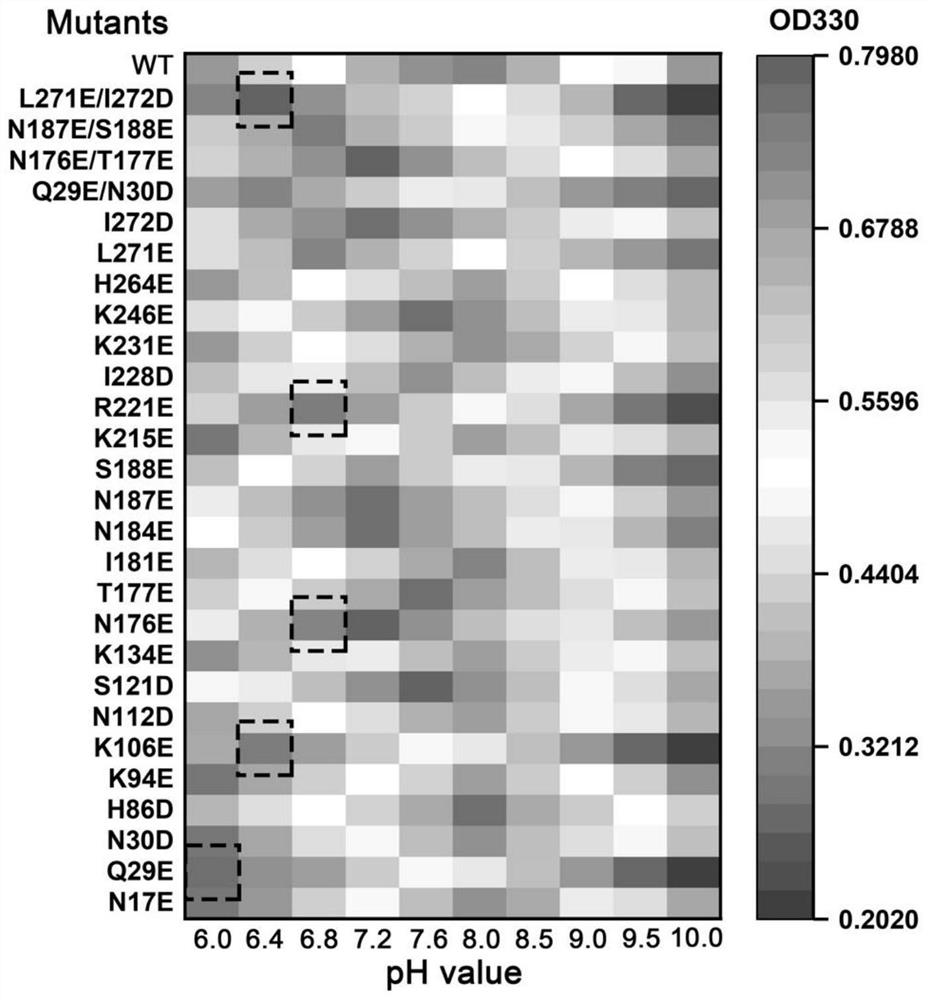
[0051] Pick a single colony and inoculate it in a sterilized 96-deep well plate containing liquid LB medium (600 μL per well) containing Kan antibiotics. Cultivate at 37° C., 900 rpm for 12 hours to obtain seed liquid. The next day, transfer 200 μL of the resulting seed solution to a new sterilized 96 deep-well plate equipped with a medium (600 μL per well). The medium composition of the new 96 deep-well plate is: Kan antibiotics, the final concentration of 0.05mmol / L IPTG LB liquid medium. At 37°C, culture was induced for 5-8 hours on a shaking shaker at 900 rpm. Use a 96-well plate centrifuge to centrifuge the 96-well plate of cultured bacteria at 3,500×g for 5 min, discard the supernatant, and add 100 μL of 50 g / L GlcNAc solution (solvent is PB buffer at pH 6.0-10.0) to each well. The bacteria were suspended and reacted at 40° C. on a shaking table at 900 rpm for 20 minutes. Then 50 μL of terminator (0.5 mol / L HCl) was add...
Embodiment 3
[0053] Embodiment 3 combined mutant specific enzyme activity detection
[0054] Combined mutations were performed on the 5 sites screened in Example 2. Using p43NMK-M14 as a template, the sequence is shown in SEQ ID NO.2. Design site-directed mutagenesis primers (as shown in Table 1) for PCR amplification and transformation of Escherichia coli JM109. After extracting the plasmid, it was transformed into B. subtilis WB600. The combined mutants and mutation sites are shown in Table 3.
[0055] Table 3 Combined mutants and mutation sites
[0056]
[0057]
[0058] The monoclonal transformants of the five combined mutant strains constructed were picked into 50 mL centrifuge tubes containing 5 mL liquid LB medium for seed culture, and 10 mg / mL kanamycin was added to each tube. The seed solution was cultured on a spring shaker at 37°C for 12 hours, and then transferred to a 500mL Erlenmeyer flask containing 96mL liquid TB medium with a 4% inoculum size for fermentation, an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Specific enzyme activity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com