Copper-containing amine oxidase sourced from saccharopolyspora cavaleriei and capable of degrading biogenic amine and application thereof
A copper amine oxidase and recombinant microorganism technology, applied in the field of molecular biology, can solve the problem of high biogenic amine content, achieve the effects of wide degradation spectrum and improved safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
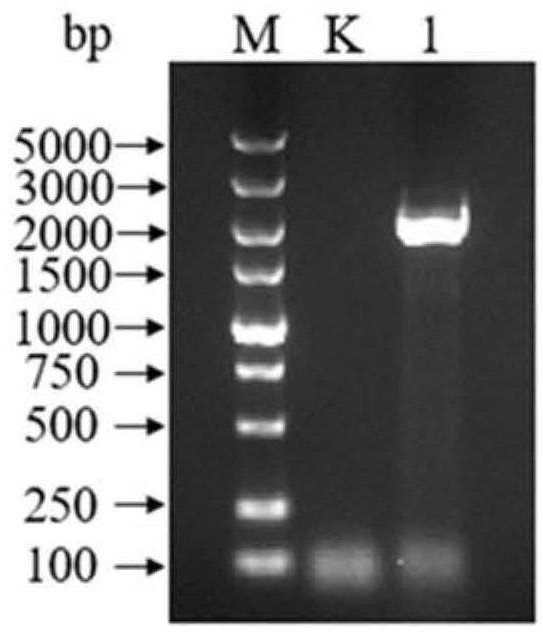
[0046] Example 1: PCR amplification of the copper-containing amine oxidase gene in S.hirsuta J2
[0047] Primers were designed according to the amine oxidase gene (Protein ID is WP_150069050.1) in Saccharopolyspora hirsuta (Saccharopolyspora hirsuta) in the NCBI database, and the S.hirsuta J2 (preservation number: CCTCC NO: M 2020103, disclosed in the patent application document with publication number CN111961615A) the genome is used as a template to amplify the multi-copper oxidase gene. The primers required for amplification are as follows: the upper primer sequence (5'→3') is ATGATGGCGATGCACCCGCTGG; the lower primer sequence (5'→3') is TCAGGACTCGCAGCAGTGGG. according to Requirements for the reaction system of HS DNA Polymerase with GC Buffer Configure the PCR reaction solution. The PCR amplification system is as follows: pre-denaturation at 98°C for 10s, annealing at 55°C for 30s, extension at 72°C (1min·kb -1 ) cycle 30 times.
[0048] Using the genome of S.hirsuta J2...
Embodiment 2
[0049] Example 2: Genetically engineered bacteria E.coli BL21-pET28a-CuAO Shir build
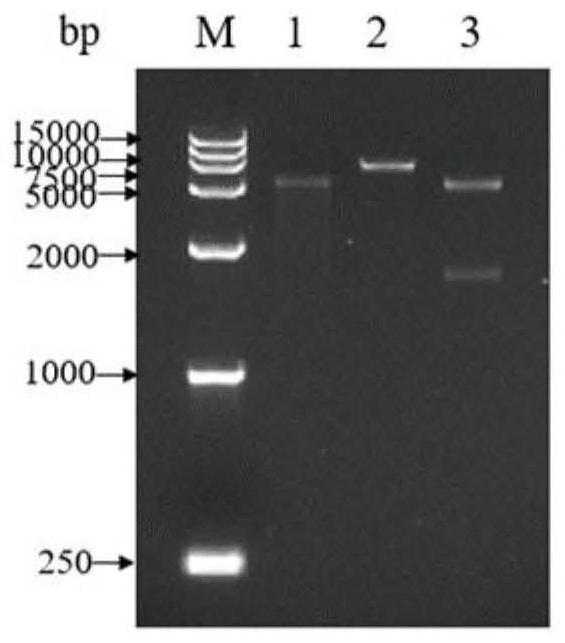
[0050] (1) Obtain the target fragment.
[0051] Using the whole genome sequence of S. hirsuta J2 as a template, the primers and the whole genome DNA are used to complete PCR amplification. The PCR reaction system and amplification procedure are the same as those described in Example 1, and the correct PCR product gel is carefully cut for band verification. , recovered and purified.
[0052] (2) Digestion and ligation.
[0053] The plasmid pET-28a(+) and the target fragment were double-digested with restriction endonuclease Nde I and EcoR I respectively. The restriction enzyme digestion system was as follows: 40 μL of target gene fragment, 40 μL of plasmid, I and EcoR I each 2.5 μL, Green Buffer 5 μL. The components in the enzymatic cleavage system were thoroughly mixed, and then placed in a metal bath at 37°C for 45 minutes of reaction. After recovery and purification, the double-digest...
Embodiment 3
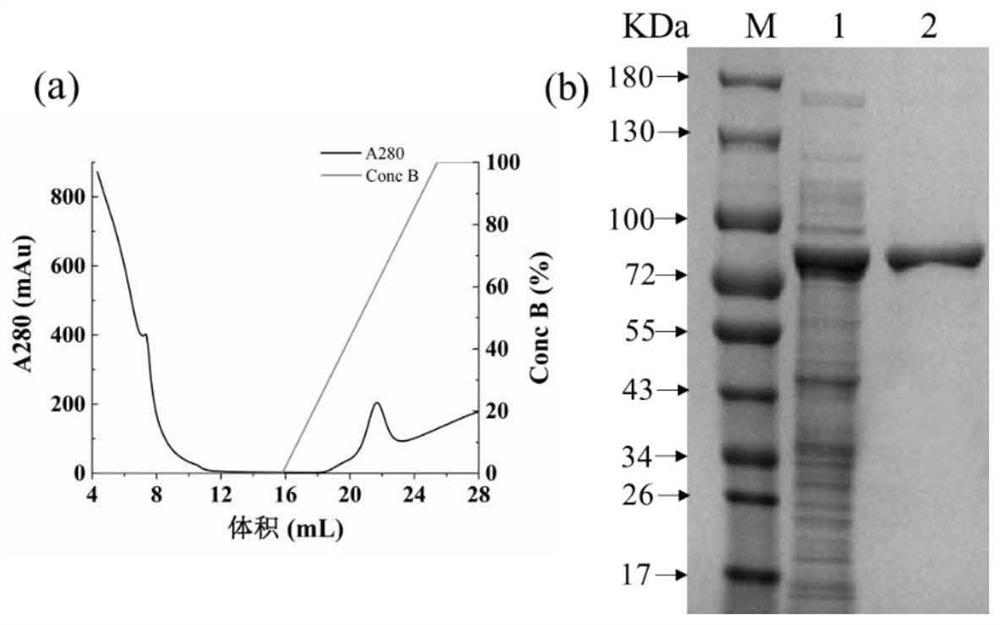
[0058] Embodiment 3: Recombinase CuAO Shir induced expression and purification
[0059] (1) Recombinase CuAO Shir induced expression of
[0060] The recombinant bacterium constructed in Example 2 was inoculated to contain 50 mg·L -1 In LB medium of ampicillin, at 37°C, 150r·min -1 Conditioned for 12h. The seed solution was transferred to the seed solution containing 50mg·L -1 In the TB fermentation medium of kanamycin, at 37°C, 160r·min -1 Grow to OD 600 is 0.6, and the final concentration is 0.25mmol L -1 IPTG, at 25°C, 160r·min -1 After culturing for 12 hours under the same conditions, the bacterial solution OD 600 is 1.5. Bacterial liquid at 4 ℃, 12000r min -1 Under these conditions, after centrifugation for 10 min, the lower layer of bacteria was collected, and 0.2 mol·L -1 The cells were resuspended in sodium phosphate buffer (pH 7.4) and collected by centrifugation, and the above steps were repeated twice. Use an ultrasonic cell disruptor to crush the bacter...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Specific vitality | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com