Camellia oleifera self-incompatibility associated gene, SNP (Single Nucleotide Polymorphism) molecular marker and application
A molecular marker, SNP181968 technology, applied in the field of molecular markers, can solve problems such as affecting the agronomic traits of Camellia oleifera
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
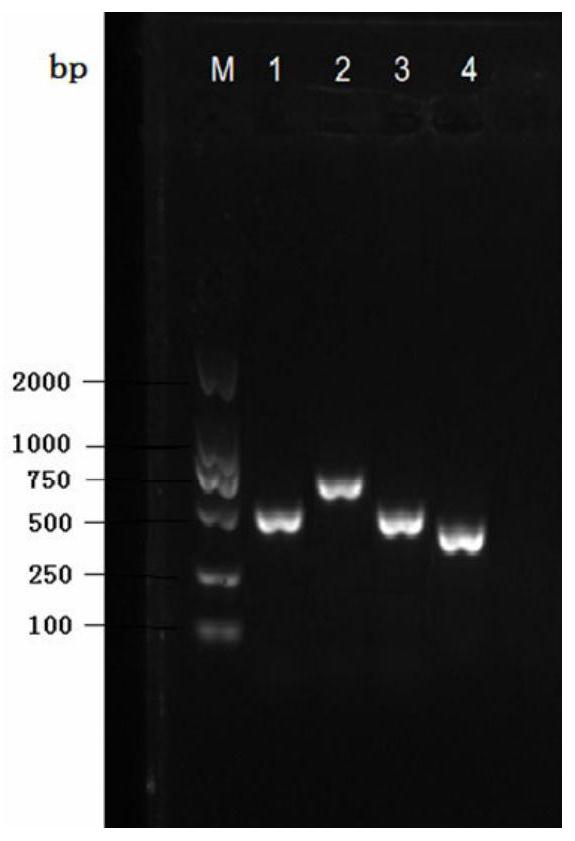
[0038] Example 1, total DNA extraction and fruit setting rate calculation of Camellia oleifera seed kernel, root or leaf
[0039] The DNA secure new plant genomic DNA extraction kit (TIANGEN Kit Code No. DP320, spin column type) combined with the improved CTAB method in the laboratory was used to extract the total DNA of a single plant. The specific operation steps are as follows:
[0040] (1) Take 600 μL of 2×CTAB solution in a 1.5mL centrifuge tube, mix well and keep the temperature in an air heater at 65°C for later use;
[0041] (2) quickly take out the camellia oleifera seeds, roots or leaves from the liquid nitrogen, and quickly grind them into fine powder in a pre-cooled mortar filled with liquid nitrogen;
[0042] (3) Transfer the ground powder to (1) the prepared centrifuge tube, mix the powder and the solution on a vortex instrument, and put it in a water bath at 65°C for 10 minutes, and shake it every 2 minutes during this period;
[0043] (4) Add 600 μL of chloro...
Embodiment 2
[0049] Example 2. Three-generation sequencing + next-generation sequencing samples and annotation analysis
[0050] 1. Sequencing sample preparation and collection:
[0051] The self-pollination combination (Self -pollination: SP), cross-pollination combination (Cross-pollination: CP) and non-pollination combination (Non-pollination: NP) ('Joajin' and 'Huaxin' emasculation treatment bagging). Cross-pollination was done manually Pollination is complete. Pick the flower buds of 'Huaxin' and 'Huajin' at the dew stage, gently stroke the anthers with tweezers, spread the anthers in a sulfuric acid paper bag, and fully crack the anthers at 25°C to release mature pollen (Pollen: Pn); store the pollen in a 1.5mL centrifuge tube at -80°C; take the pollen out of the refrigerator and equilibrate it at room temperature for 1 hour before pollination. When pollinating, select flower buds in the white stage (after the flower buds are peeled off, the anthers are not powdered and the pistil i...
Embodiment 3
[0059] Example 3: CoMAPK gene acquisition and SNP site analysis
[0060] 1. Analysis of CoMAPK9 gene characteristics related to self-incompatibility
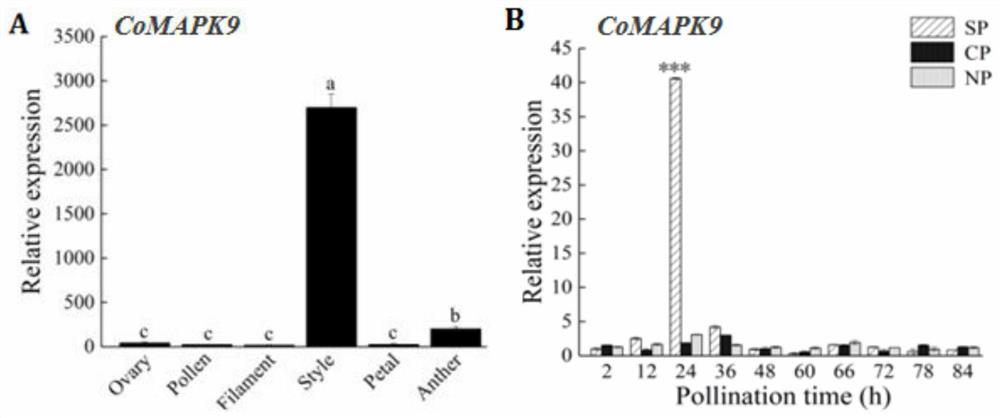
[0061] Transcript annotation analysis, differential transcripts, and differential protein trend analysis and Pearson correlation analysis were performed, and CoMAPK9 was found to be a unique sequence of Camellia oleifera, which strongly responded to self-pollination and out-pollination. The functional characteristics of CoMAPK9 gene in the process of Camellia oleifera self-incompatibility were studied by sequence analysis, expression pattern analysis, prokaryotic expression, subcellular localization and other experiments.
[0062] Using real-time fluorescent quantitative PCR to analyze the expression of CoMAPK9 in different parts of flowers ( figure 2 A), the expression patterns of 2-84h in the styles of different treatments ( figure 2 B).
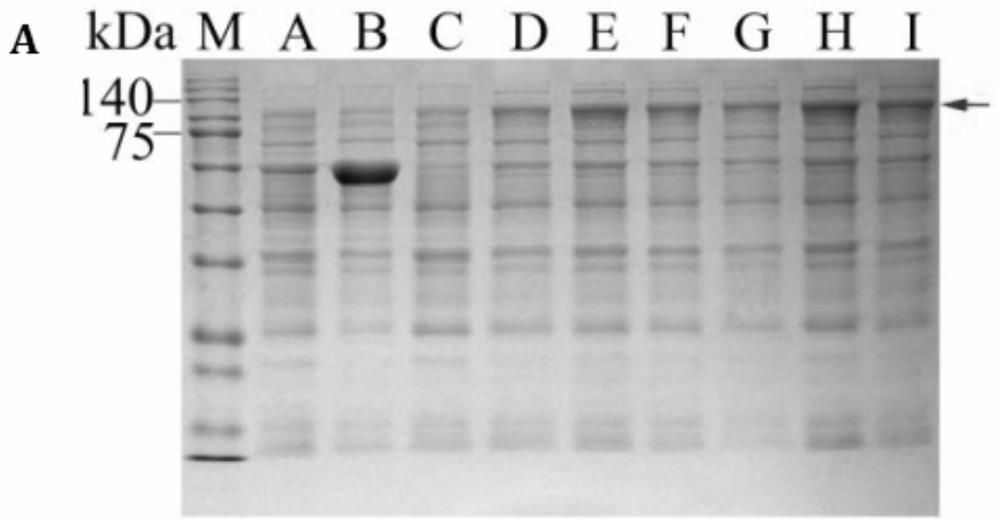
[0063] CoMAPK9 protein expression was induced by prokaryotic expression experiments...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com