Fluorescent quantitative PCR (polymerase chain reaction) detection method and kit for plant pathogenic bacterium pantoea ananatis
A detection kit, the technology of Pantoea pineapple, is applied in the field of molecular biology to achieve the effect of a highly specific and accurate detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
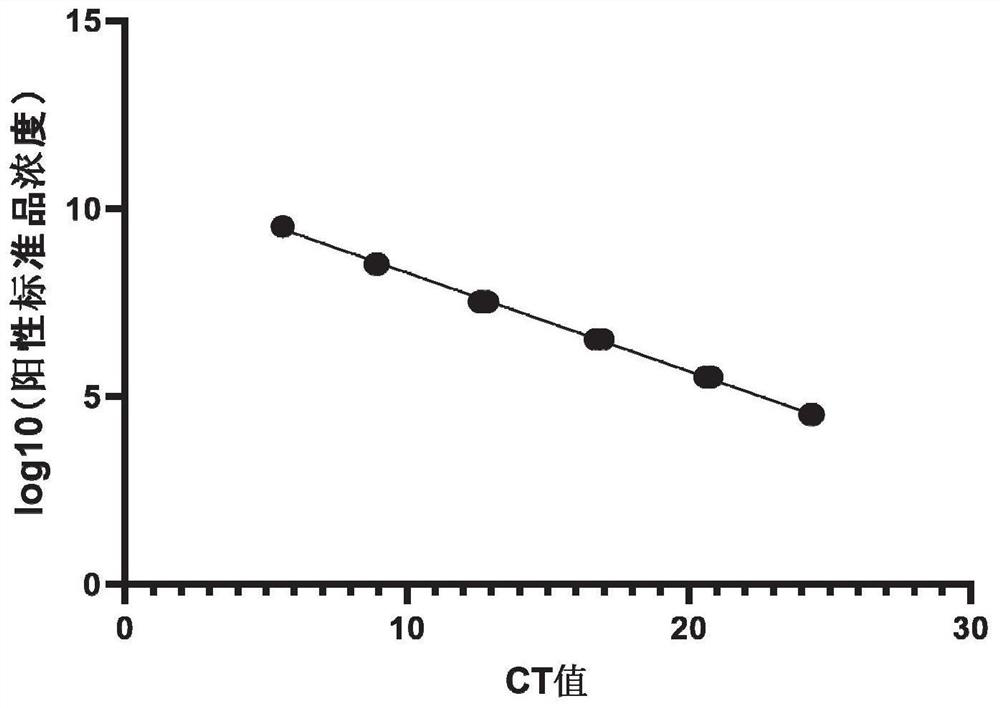
[0026] The preparation process of the positive standard gradient is as follows: take 10 μl of the positive standard plasmid, dilute it in a 10-fold gradient, and sequentially dilute 4-7 gradients to obtain the positive standard gradient.
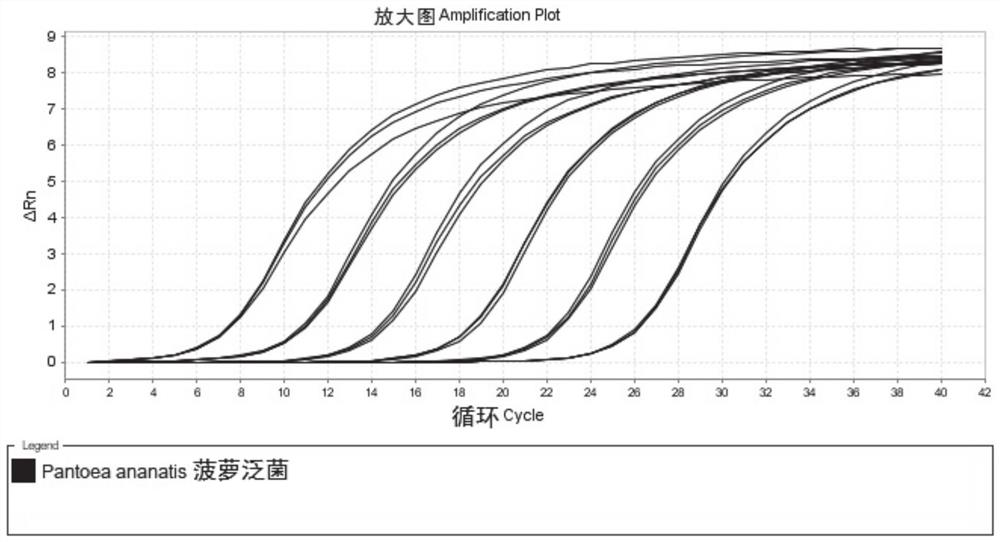
[0027] After the gradient preparation of the positive standard is completed, the fluorescence quantitative PCR reaction is carried out under the same conditions with the gradient positive standard and sample DNA, and the concentration logarithm of the gradient positive standard is taken as the ordinate, and the Ct value of the experimental result is drawn as the abscissa. , according to the Ct value obtained by the sample test, calculate the concentration of Pantoea pineapple in the sample.
[0028] The present invention also provides a method for detecting the fluorescent quantitative PCR of the plant pathogenic bacterium Pantoea pineapple by the kit, the method comprising: step 1, extracting the genomic DNA of the sample to be tested; step ...
Embodiment 1
[0033] 1. Design of specific primers for detecting Pantoea pineapple.
[0034] Specific primers were designed and synthesized for Pantoea pineapple gyrB gene sequence (GenBank accession number: MW981333.1), and the designed specific primer sequences were analyzed by BLAST. The results showed that the primers had high specificity. Its sequence is as follows:
[0035] Upstream primer: TATCCGCGCCTTTGTTGAGT (SEQ ID NO: 1).
[0036] Downstream primer: ATGCCGTCTTTCTCGGTTGA (SEQ ID NO: 2).
[0037] Design and synthesize positive standard sequences:
[0038] 5- TATCCGCGCCTTTGTTGAGTACCTGAATAAAAACAAAACGCCTATTCACCCAACCGTATTTCTATTTCTCAACCGAGAAAGACGGCAT-3 (SEQ ID NO: 3).
[0039] 2. Extract the genomic DNA of the sample to be tested
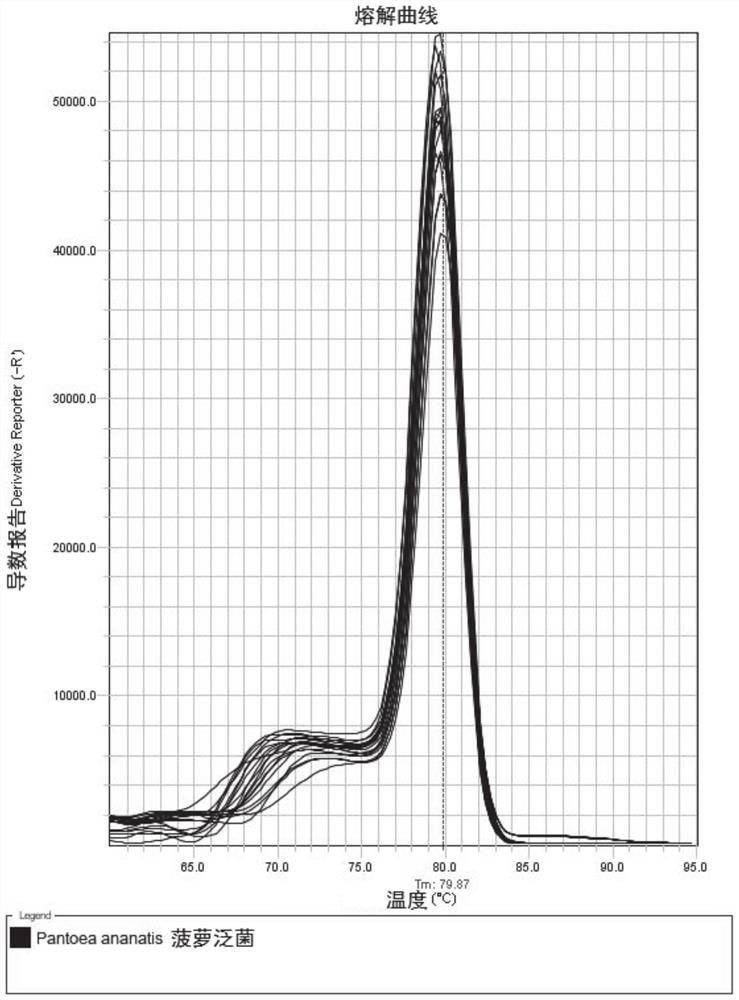
[0040] 3. Using the extracted genomic DNA as a template, set a positive standard control and a negative control, prepare a PCR reaction system with the Pantoea pineapple PCR reaction solution and specific primers, and perform a fluorescent quantitative PC...
Embodiment 2
[0049] Example 2. Performance evaluation of fluorescent quantitative PCR detection method.
[0050] 1. Sensitivity assessment of the detection method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com