Dna-cutting agent
A technology of DNA molecules and DNA sequences, which is applied in the fields of molecular biology and microbiology, can solve problems such as inability to develop and reduce acetic acid production, and achieve the effect of increasing versatility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1. Testing the activity of СсCas9 protein in cleaving various DNA targets
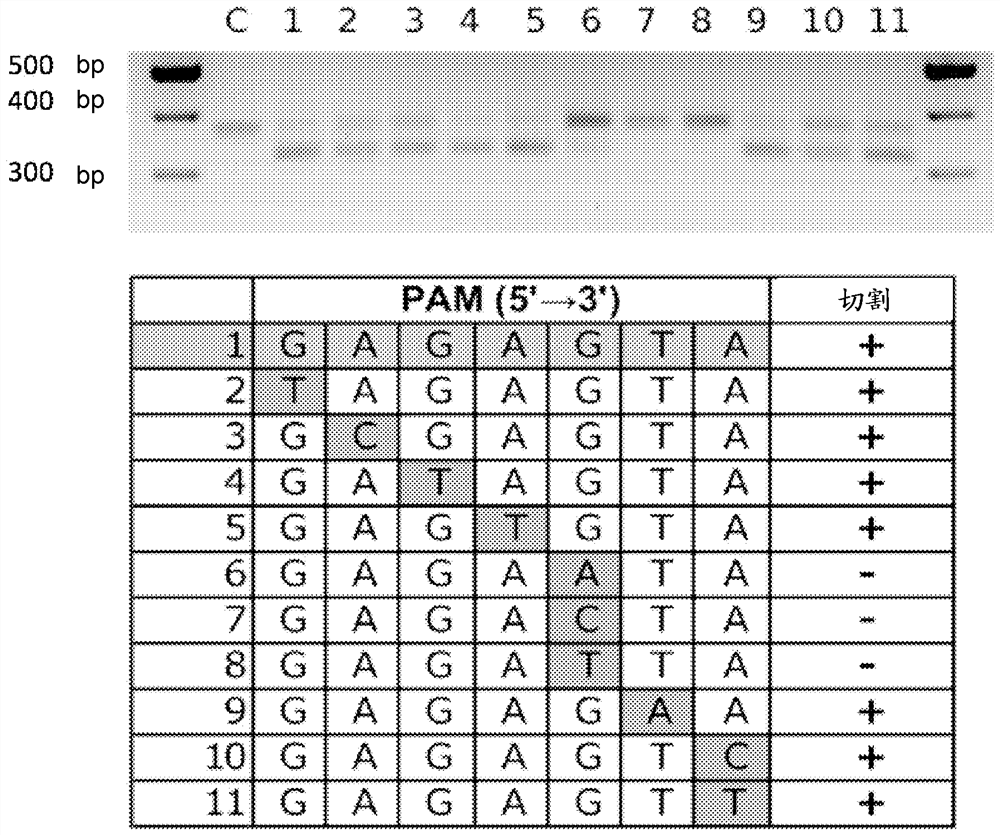
[0074] To examine the ability of CcCas9 to recognize various DNA sequences flanked by NNNNGNA 3' sequences, experiments were performed on in vitro cleavage of DNA targets from the human grin2b gene sequence (see Table 1 below).
[0075] Table 1. DNA targets isolated from the human grin2b gene.
[0076] DNA target PAM ctacatcacgtaacctgtct tagaAgA gaacgagctctgctgcctga cacgGcc agaacgagctctgctgcctg acacGgc acggccaaccaccaaccagaa cttgGgA tccgctctgggcttcatctt caactcg cgactccctgcaaacacaaa gaaagag atctacatcacgtaacctgt cttaGaA tatctcctttcattgagcac caaaccc
[0077] Perform in vitro DNA cleavage reactions under similar conditions to the experiments described above. As a DNA target, a fragment of the human grin2b gene with a size of about 500bp was used:
[0078]
[0079] Experimental results show that CcCas9 in complex with...
Embodiment 2
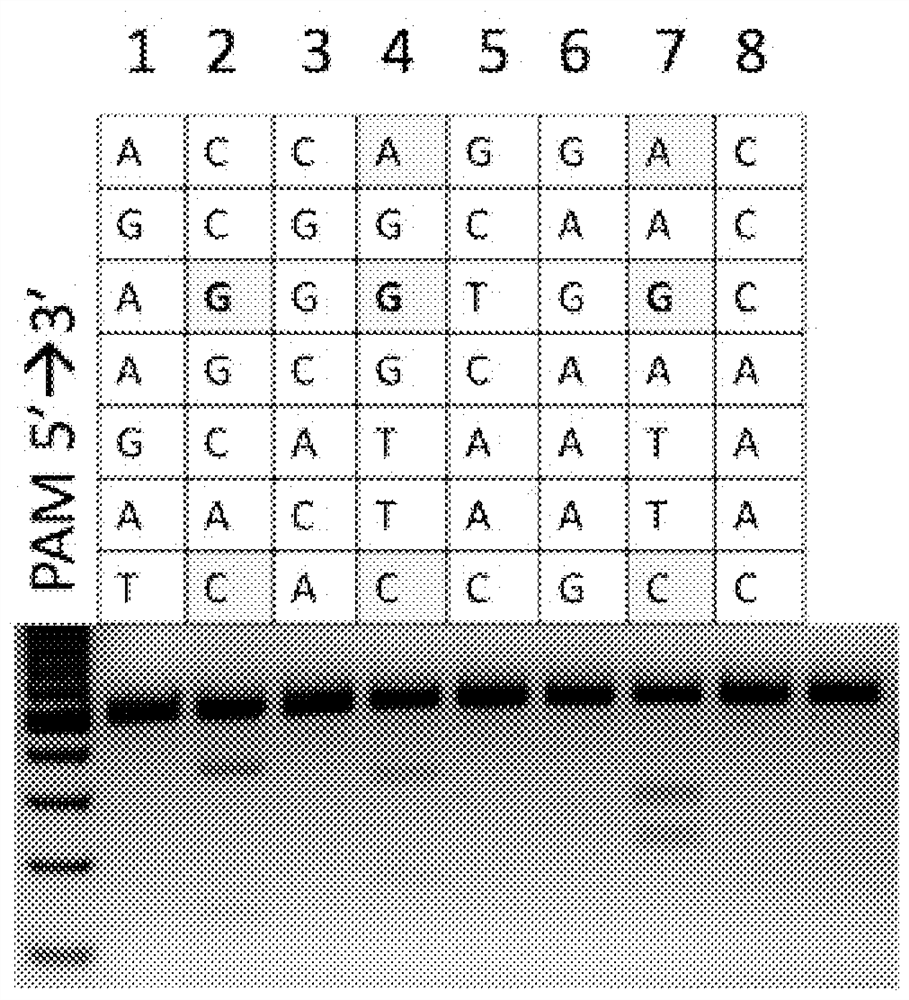
[0080] Example 2. Temperature range of CcCas9 activity
[0081] In order to determine the temperature range of the CcCas9 protein, experiments on in vitro cleavage of DNA targets were performed under different temperature conditions.
[0082] To this end, target DNA flanked by the PAM sequence GAGAGTA was subjected to cleavage at different temperatures by the CcCas9 effector complex with the corresponding guide RNA ( Figure 6 ).
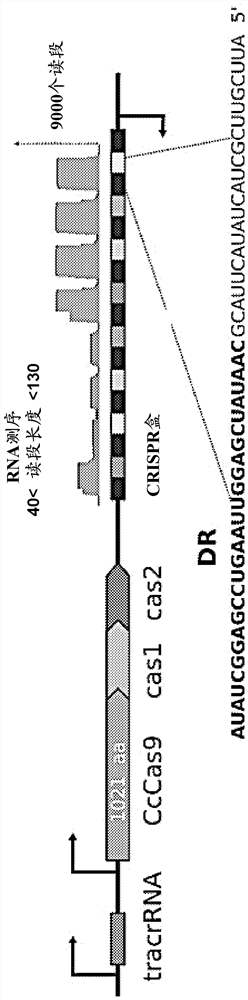
[0083] The CcCas9 protein was found to have activity over a wide temperature range. Maximum nuclease activity is achieved at a temperature of 45°C, while the protein is sufficiently active in the range of 37°C to 55°C. Thus, CcCas9 in complex with guide RNA is a new tool for cleavage (double-strand break formation) in DNA molecules restricted to the sequence 5'-NNNNNGA-3', with a temperature range of 37°C to 55°C. A schematic diagram of the complex of target DNA with crRNA and tracer RNA (tracrRNA) that together form a guide RNA is shown in Fig...
Embodiment 3
[0085] from the genus Clostridium ( Clostridium ) of the closely related organism Cas9 protein. So far, only one type II CRISPR Cas system has been found in Clostridium, which is derived from Clostridium perfringens ( Clostridium perfringens) Cas9 CRISPR Cas system (Maikova A et al., New Insights Into Functions and Possible Applications of Clostridium difficile CRISPR-Cas System .Front Microbiol. 2018 Jul 31;9:1740).
[0086] The Cas9 protein from Clostridium perfringens bacteria has 36% identity with the CcCas9 protein (the degree of identity is calculated using BLASTp software, default parameters). A comparable Cas9 protein from S. aureus in size has 28% identity to CcCas9 (BLASTp, default parameters).
[0087] Thus, the CcCas9 protein differs significantly in amino acid sequence from other Cas9 proteins studied to date, including Cas9 proteins found in related organisms.
[0088] Those skilled in the field of genetic engineering will understand that the CcCas9 protei...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com