Application of nitrosation modification of transcriptional regulatory factors WalR and MgrA in treatment of staphylococcus aureus infection
A staphylococcus infection, nitrosylation technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] Construction of embodiment 1.nos knockout strain:
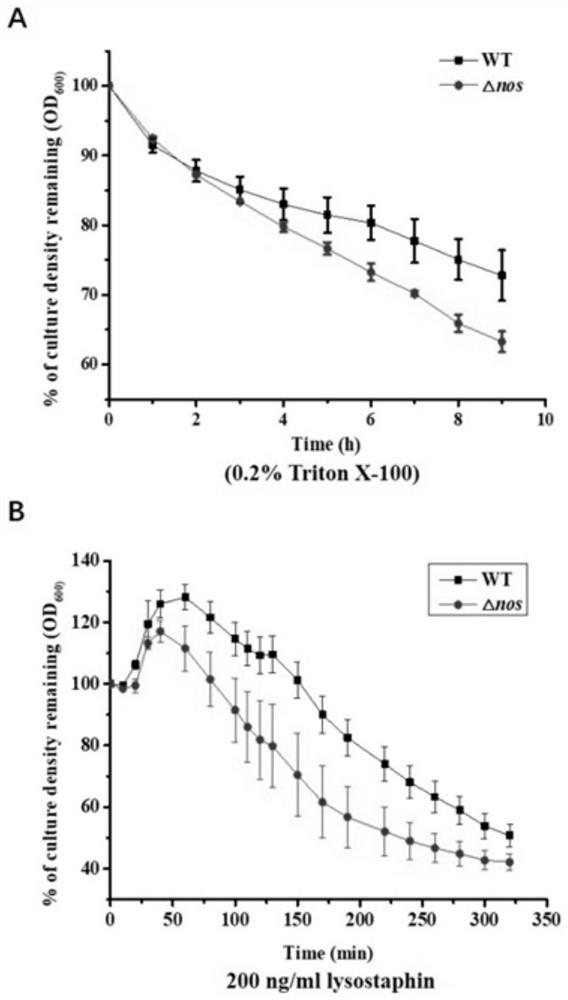
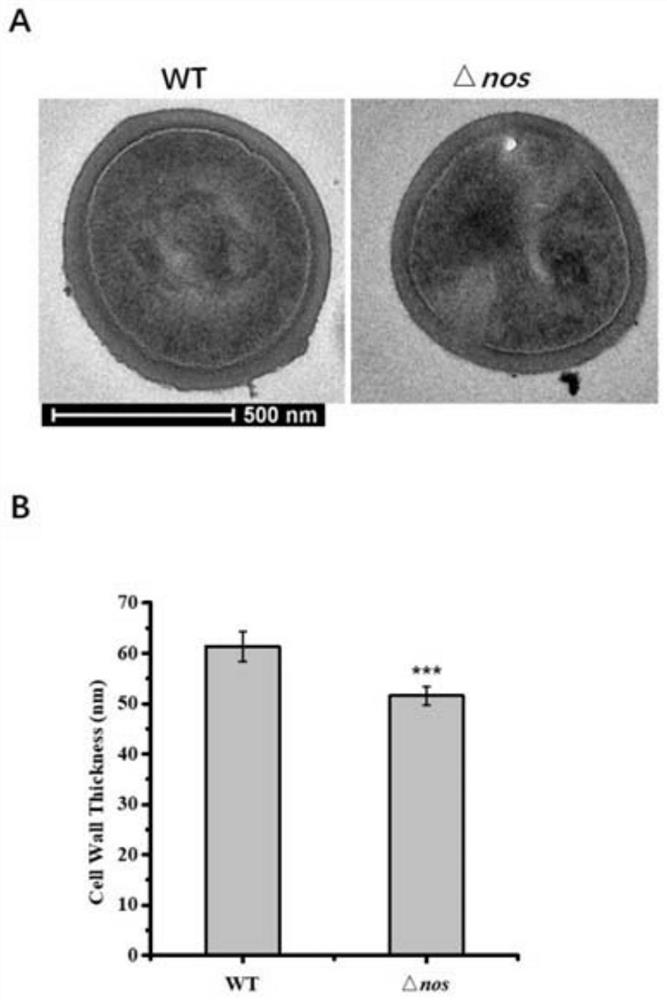
[0095] The nos gene is used to encode the NOS protein (nitric oxide synthase), which produces NO, and then mediates the nitrosylation modification of the cysteine site of the protein.
[0096] In this example, the target gene traceless knockout technology is adopted, and the homologous recombination plasmid is constructed with the temperature-sensitive plasmid pBTs as the carrier. The method is as follows: use HS DNA Polymerase (TaKaRa, DR010A) amplifies the upstream and downstream homology arm fragments of the nnos gene with bNOS-up-F and bNOS-up-R, bNOS-down-F and bNOS-down-R from the XN108 genome, respectively, by PCR was performed to overlap the upstream and downstream to obtain combined fragments, and the corresponding fast restriction endonucleases KpnI (Thermo, FD0524) and SacI (Thermo, FD1133) were used to treat the vector pBTs and fragments respectively, and after recovery and purification, T4 DNA ligase (T...
Embodiment 2
[0108] Example 2. Identification of protein targets that can be modified by bacterial endogenous NO nitrosylation:
[0109] In this example, iodoTMT (irreversible isobaric iodoacetyl Cys-reactivetandem mass tag) is used to carry out isotope labeling and enrichment of proteins modified by SNO nitrosylation, and then combined with mass spectrometry-based proteomics analysis, and nos knockout strains were combined with Wild-type strains were compared to identify protein targets in S. aureus that could be modified by nitrosylation by endogenous bacterial NO. iodoTMT markers (thermo, 90100, 90101, 90102, 90103) and proteomics were completed by Jingjie Biological Company.
Embodiment 3
[0110] Example 3. Construction of amino acid point mutation strains:
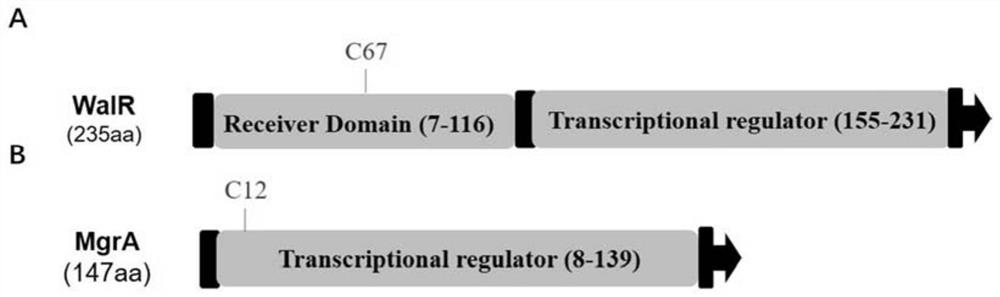
[0111] The walR (C67S) and mgrA (C12S) single-point mutant strains in the strain XN108 were constructed using the target gene seamless knockout technology, and the temperature-sensitive plasmid pBTs was used as a carrier to construct a homologous recombination plasmid. The method is as follows:
[0112] use HS DNA Polymerase (TaKaRa, DR010A) amplifies the walR gene from the XN108 genome with walR-C67S-up-F and walR-C67S-up-R, walR-C67S-down-F and walR-C67S-down-R, respectively The 67th cysteine (C67) upstream and downstream homology arm fragment; use mgrA-C12S-up-F and mgrA-C12S-up-R, mgrA-C12S-down-F and mgrA-C12S-down-R respectively Amplify the homology arm fragments upstream and downstream of the 12th cysteine (C12) of the mgrA gene. The combined fragments were obtained by overlapping the upstream and downstream by PCR respectively. The upstream and downstream combined fragments of walR (C67S) we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com