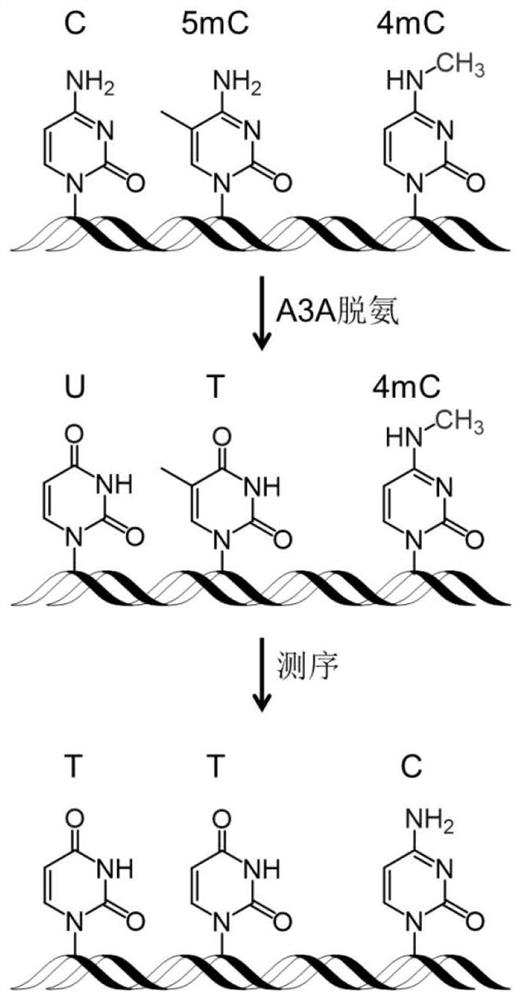
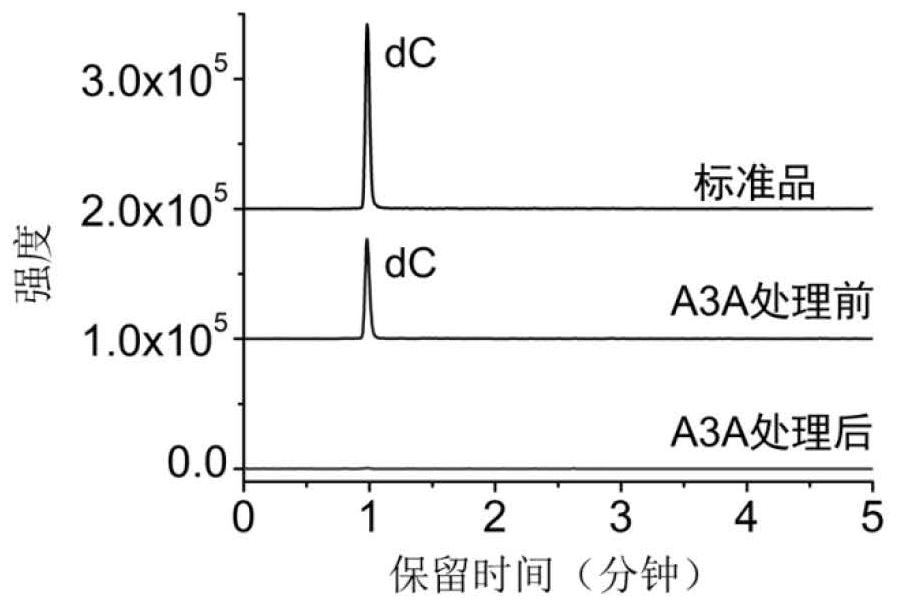
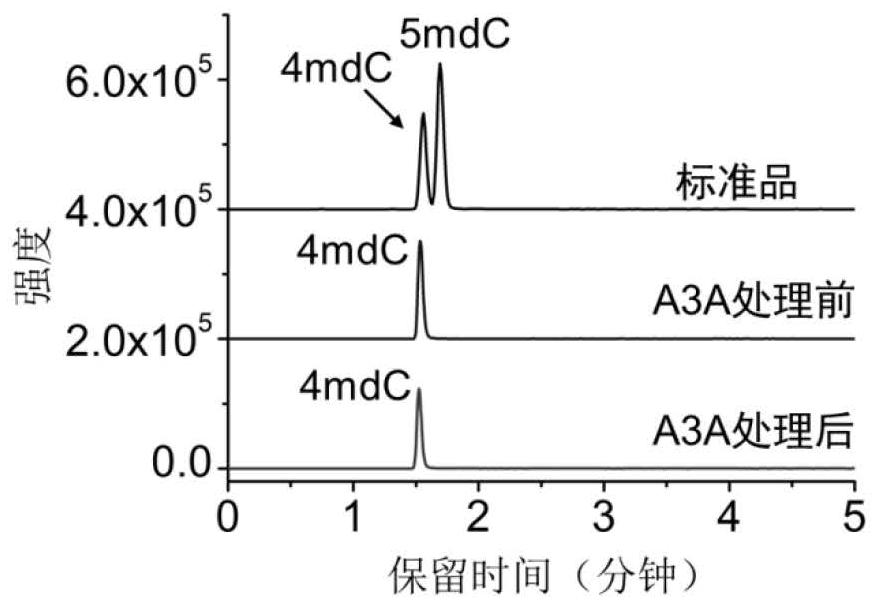
Single-base resolution positioning analysis method for N4-methylcytosine in deaminase-mediated DNA
A methylcytosine and analysis method technology, applied in the field of single-base resolution positioning analysis of N4-methylcytosine, to achieve the effects of shortened analysis time, mild reaction conditions, and small dosage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1. Utilize artificially synthesized 215bp DNA strands containing different modifications to distinguish cytosine, 5-methylcytosine and N 4 - Methylcytosine
[0049] In order to verify the accuracy of the method of the present invention, a 215bp DNA chain containing different modifications was first synthesized, reacted with A3A protein, and the deamination rate was verified by mass spectrometry and Sanger sequencing.
[0050] A. Synthetic method of 215bp DNA chain
[0051] In a 50 μL reaction system, add 0.5ng pUC19 DNA (Takara Biotechnology Co., Ltd.), 5 μL 10× reaction buffer, 2.5U Taq DNA polymerase (Takara), 2 μL 10 μM forward primer (sequence 5′-GAGTGAGTGAGGGAGGAAG -3'), 2 μL of 10 μM reverse primer (sequence 5'-CCACTCACAATTCCACACAACATAC-3'), 1 μL of dATP, dGTP, TTP and dCTP (all at a concentration of 2.5 mM). When synthesizing 5mC or 4mC modified DNA strands, just replace dCTP with 5mdCTP or 4mdCTP.
[0052] The polymerase chain reaction (PCR) progra...
Embodiment 2
[0062] Example 2. The deaminase-mediated 4mC single-base mapping method in DNA is used for the identification of the 4mC site in the whole genome of Deinococcus radiodurans
[0063] Deinococcus radiodurans is a specialized bacterium that is extremely resistant to the deadly effects of ionizing radiation, UV light, oxidation and desiccation. Studies have shown that 4mC plays an important role in maintaining the genome stability of Deinococcus radiodurans. Current methods for identifying 4mC are mainly based on bisulfite conversion or third-generation sequencing technologies, however, each of these technologies has its own shortcomings. For example, bisulfite conversion can cause a lot of degradation to DNA, and the accuracy of third-generation sequencing has always been a problem. Therefore, using the method established above, the 4mC modification at a specific site in the genomic DNA of Deinococcus radiodurans was identified.
[0064] In order to explore the distribution and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com