Method for rapidly obtaining probiotic drug-resistant gene in sterilized dairy product based on high-throughput sequencing technology
A technology for drug resistance genes and dairy products, applied in the field of biological detection, can solve problems such as inability to achieve detection, and achieve the effect of being suitable for large-scale sample extraction and cheap.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 sample total DNA preparation
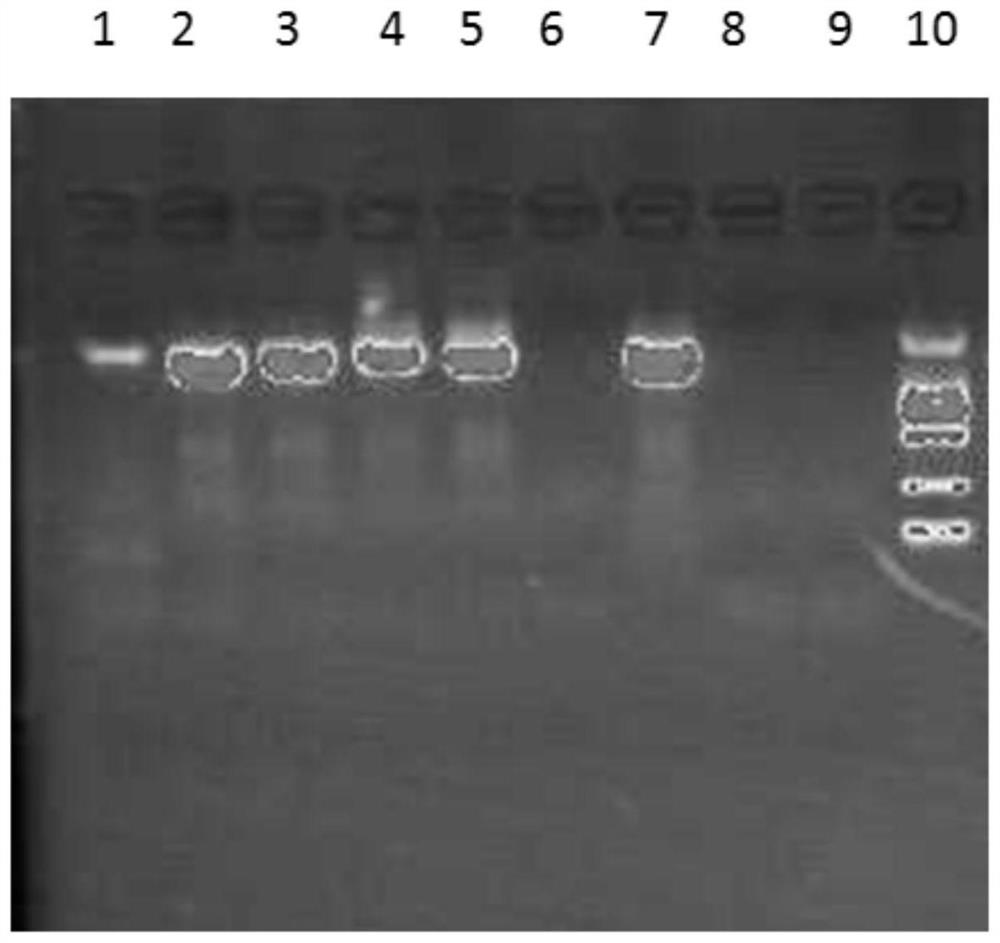
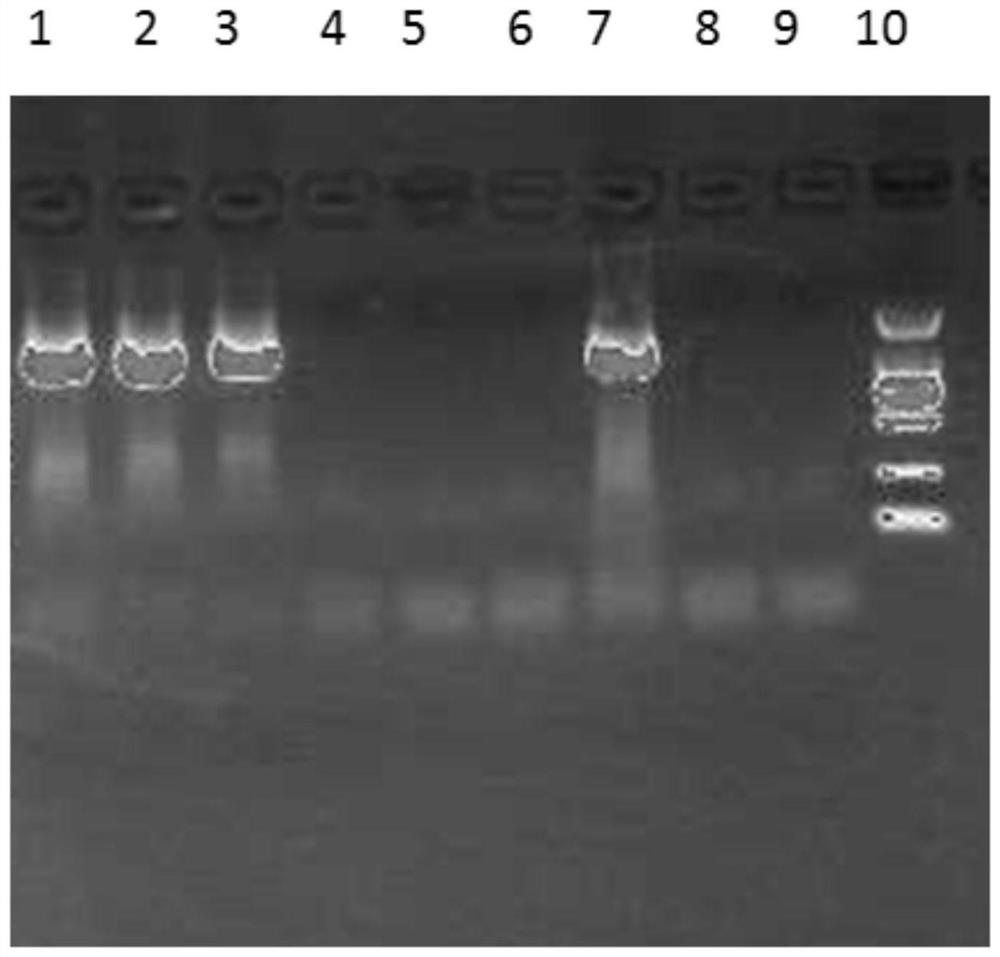
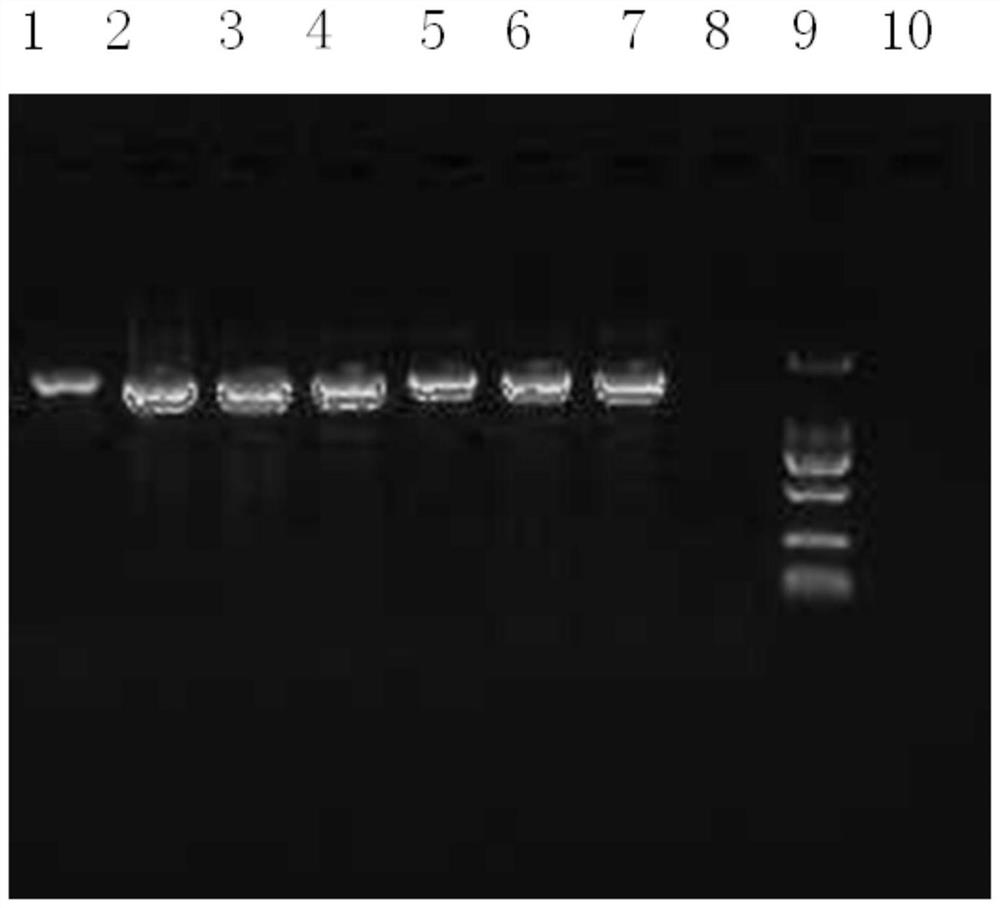
[0042] 1. The quality of bacterial nucleic acid extraction will directly affect subsequent experiments. It is necessary to consider the particularity of the samples to be tested and the rich diversity of bacterial species. DNA extraction is the basis for subsequent amplification, sequencing and analysis. How to extract samples efficiently and comprehensively The desired DNA in , has a particularly prominent impact on the results. At present, there are various methods of DNA extraction with different principles and each has its own advantages and disadvantages. In this example, 6 representative examples of dairy products with bactericidal probiotics in the market were selected, including 3 examples of fermented milk and 3 examples of milk-containing beverages. The deep-processed food DNA extraction kit method, the magnetic bead method universal genomic DNA extraction kit method and the applicant's improved CTAB method were selected...
Embodiment 2
[0094] Example 2 Using high-throughput sequencing to obtain the sequence of the template DNA fragment
[0095] 1. Fragmentation of the DNA sample extracted in Example 1: about 350bp
[0096] Instrument: Covaris M220
[0097] 2. Construction of PE library
[0098] (1) Connect the "Y" shaped connector;
[0099] (2) Use magnetic beads to screen and remove adapter self-ligation fragments;
[0100] (3) Enrichment of library templates by PCR amplification;
[0101] (4) Sodium hydroxide denatures to produce single-stranded DNA fragments.
[0102] Reagents: TruSeq TM DNA Sample Prep Kit
[0103] 3. Bridge PCR
[0104] (1) One end of the above-mentioned single-stranded DNA fragment is complementary to the base of the primer, and fixed on the chip;
[0105] (2) The other end is randomly complementary to another nearby primer and is also fixed to form a "bridge";
[0106] (3) PCR amplification to generate DNA clusters;
[0107] (4) The DNA amplicon is linearized into a single ...
Embodiment 3
[0115] Example 3 Sequencing sequence statistics and quality control
[0116] (1) Raw sequence statistics
[0117] In the sequencing experiment, multiple DNA samples of Example 2 were used for parallel sequencing, and the sequences in each sample were introduced with an Index tag sequence indicating the source information of the sample. The data of each sample is distinguished according to the Index sequence, and the extracted data is saved in fastq format. In the data results of the MP or PE library, each sample has two files, fq1 and fq2, which contain the reads at both ends of the sequencing, and the order of the reads is strictly consistent and corresponding to each other.
[0118] faSTQ is a text format for storing biological sequences and corresponding quality values in Solexa sequencing technology. Each read contains 4 lines of information, of which the first and third lines are composed of the file identification mark and the read section name (ID) (the first line s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com