Soybean aphid-resistant gene as well as molecular marker and application thereof
A molecular marker and anti-aphid technology, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of low efficiency, long cycle, difficult breeding, etc., and achieve the effects of improving screening efficiency, shortening the breeding period, and high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Population construction and phenotyping
[0061] 1. Raw materials: Founder's moss soybean (sourced from the National Crop Germplasm Bank, unified number ZDD00236), judged as aphid resistance level 1 according to the resistance grading standard in Table 2, and the resistance does not depend on the number of soybean aphids.
[0062] Table 2 Grading standards for identification and grading of resistance to soybean aphids
[0063]
[0064]
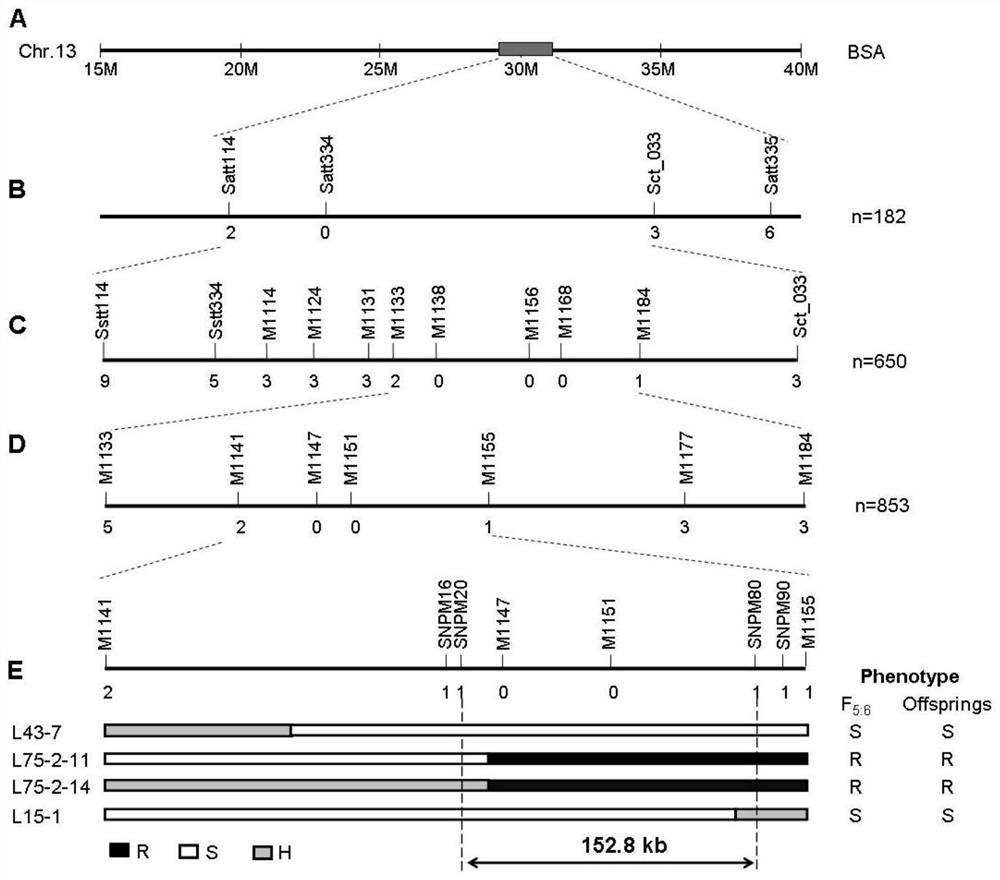
[0065] 2. Use Fangzheng moss-eating bean to construct a population, and locate the aphid-resistant gene by map-based cloning: use Fangzheng moss-eating bean and the highly susceptible aphid variety Beifeng No. 9 to cross ( figure 1 ), and then self-crossed to construct a genetic segregation population resistant to susceptible aphids. The results of resistance identification showed that F 1 Plants showed resistance to aphids, F 2 The segregation ratio of aphid-resistant plants and aphid-susceptible plants in the generation popula...
Embodiment 2
[0067] Localization of resistance genes and determination of linked molecular markers
[0068] Using the BSA (cluster segregation analysis) method, the DNA of aphid-resistant and susceptible plants was mixed to construct a mixed pool, and the 580 pairs of SSR primers covering the whole soybean genome in the SoyBase database were screened to have polymorphisms between parents and meet extreme mixed pools According to regular primers, three markers, Satt114, Sct_033 and Satt_335, were found to be resistance-linked markers. Use these 3 linked markers and encrypt other markers to identify F 2 Generation plants and by F 2 F 5 The genotype of the generation plants was combined with the phenotype for initial mapping, and the initial mapping interval was between markers M1133 and M1184, with a total of 1.01Mb. In order to further narrow the interval, the encryption mark identifies F 5:6 The genotype of the generation plant, combined with the phenotype data, screened the exchanged ...
Embodiment 3
[0075] Validation of molecular markers in progeny populations
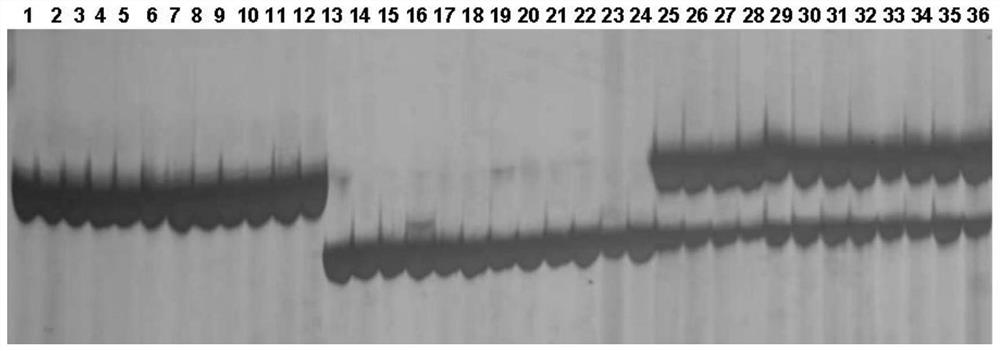
[0076] Genomic DNA from the leaves of Fangzheng Moshidou, Beifeng 9 and the offspring of the group were extracted by CTAB method and PCR amplified. The system is 20μl, including 1μl of genomic DNA, the concentration is 2mmol·L -1 2 μl each of the upstream and downstream primers, ddH 2 O 4 μl, 2хM5 HiPerplus Taq HiFi PCR Mix (with blue dye) 10 μl (Beijing Jumei Biotechnology Co., Ltd., Cat. No. MF002-plus-01). The PCR program was denaturation at 95°C for 3 min, denaturation at 95°C for 25 s, annealing at 58°C for 25 s, extension at 72°C for 10 s, and 34 cycles; extension at 72°C for 5 min, and finally storage at 4°C. The above PCR reaction was amplified on a PCR thermal cycler from ABI (Applied Biosystems, USA).
[0077] The amplified sequences labeled SNPM20 and SNPM80 were detected by 1% agarose gel electrophoresis. After obtaining a band consistent with the size of the target band, the PCR product was sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com