Specific antibody of novel coronavirus S protein as well as preparation method and application thereof
A specific, protein-based technology, applied in the direction of antiviral immunoglobulin, virus/bacteriophage, and microbial-based methods, can solve the problems of no practical clinical treatment value, large-scale mass production of high finished products, unfavorable large-scale mass production, etc. , to achieve the effects that are conducive to large-scale popularization and application, large-scale production, and large-scale clinical therapeutic use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Example 1: Preparation of antigens
[0127] The conventional practice in the prior art is to recombinantly express and purify a certain region of the S protein as an antigen.
[0128] The present invention is to prepare antigen for nearly full-length new coronavirus S protein, and the specific method is as follows:
[0129] (1) Synthesize the DNA sequence encoding the S protein (amino acids 1-1208), connect it in series with a DNA sequence encoding the T4 fibritin region, and construct it into the pFASTBAC insect cell recombinant expression vector to form the S protein trimer recombinant expression plasmid ;
[0130] The amino acid sequence of the T4 fibritin region can induce the entire peptide chain to spontaneously trimerize, and then fold to form an S protein trimer;
[0131] The amino acid sequence, nucleotide sequence of the above-mentioned S protein and the amino acid sequence of the T4 fibritin region are shown in Table 3;
[0132] table 3
[0133]
[013...
Embodiment 2
[0145] Example 2: Alpaca immunization injection
[0146] In this example, alpacas were immunized with the antigen prepared in Example 1. Specific steps are as follows:
[0147] (1) The antigens prepared in Example 1 were equally divided into 4 parts, each of about 1 mg;
[0148] (2) The alpacas were immunized 4 times in total, and the antigens were subcutaneously injected into the animals. The first immunization was recorded as the first day, and the subsequent immunizations were on the 10th, 19th, and 28th days respectively; On the 28th day, before the fourth immunization injection, about 200 mL of alpaca venous peripheral blood was collected, and on the 42nd day, that is, 14 days after the fourth immunization, about 300 mL of alpaca venous peripheral blood was collected.
[0149] Compared with the traditional immunization technical solutions for animal antibodies such as mice and rabbits, the method provided in this example collects a large amount of alpaca venous peripher...
Embodiment 3
[0150] Example 3: Construction of Antibody Libraries
[0151] Using the two batches of alpaca venous peripheral blood collected in Example 2 as raw materials, a high diversity Nanobody library was constructed. The two batches of alpaca venous peripheral blood were treated in the same way, specifically:
[0152] (1) Using density gradient centrifugation to separate lymphocytes from the peripheral blood of alpaca veins;
[0153] (2) Extracting the total mRNA of lymphocytes and reverse transcribing into cDNA;
[0154] (3) using appropriate DNA primers, using the above-mentioned cDNA as a template, through polymerase chain reaction (PCR) amplification to obtain the VHH fragments of alpaca immunoglobulin IgG2 and IgG3, that is, the DNA fragments of Nanobodies;
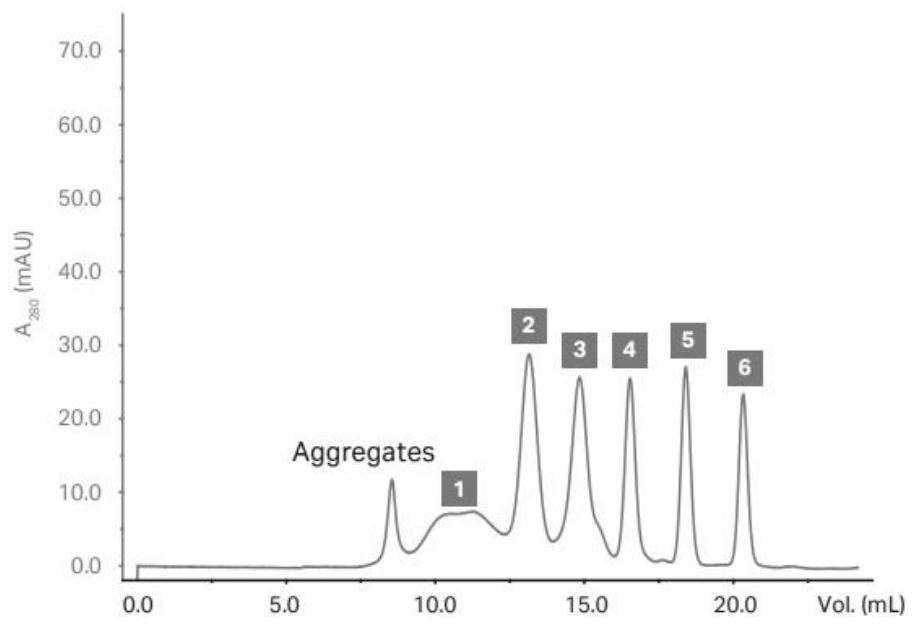
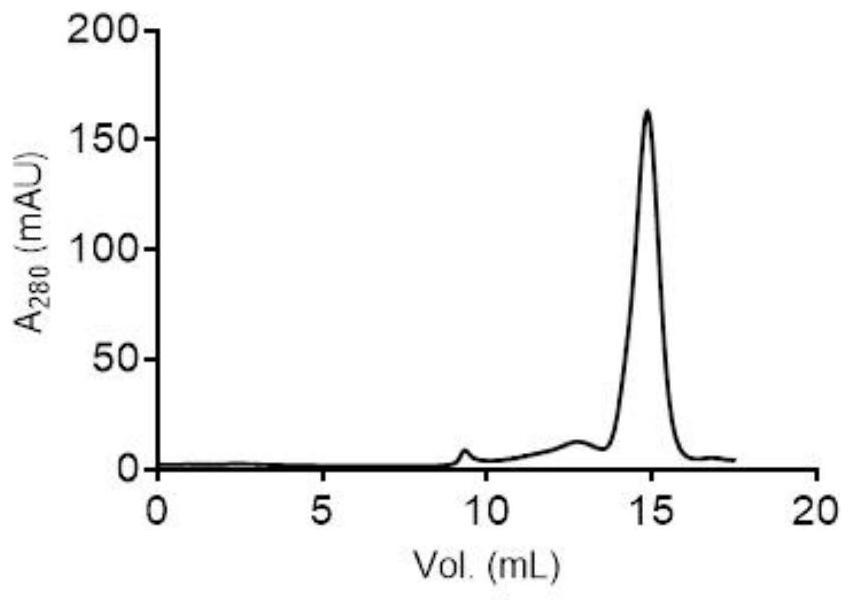
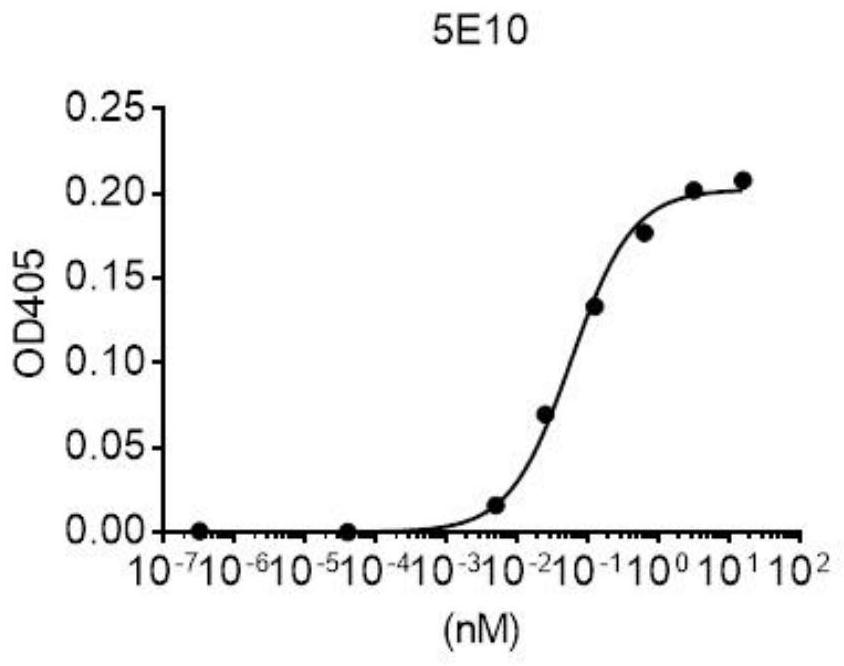
[0155] (4) the DNA of VHH is connected to the phage surface display screening vector, constitutes the VHH-pIII fusion protein expression vector plasmid library; wherein, pill is the protein present on the phage surface fl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com