High-performance k-mer frequency counting method and system based on clustering algorithm
A technology of clustering algorithm and counting method, which is applied in the field of bioinformatics, can solve the problems of low efficiency of Hash algorithm and waste memory overhead of GPU, and achieve the effect of avoiding insufficient memory, facilitating GPU calculation, and improving parallel speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
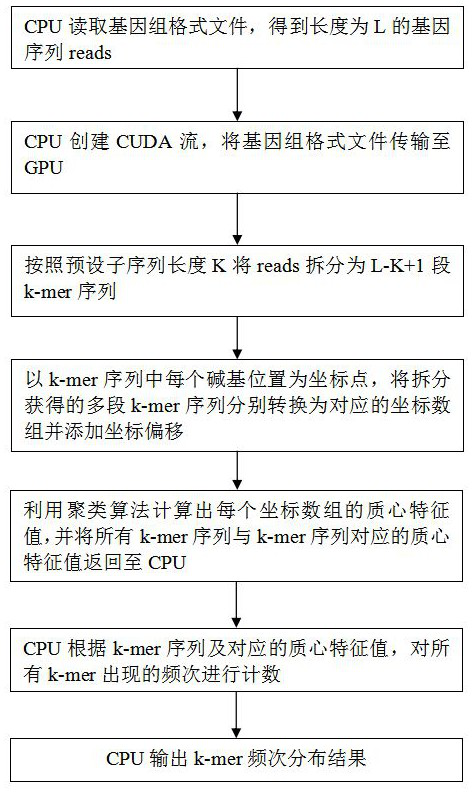
[0041] In this embodiment, as figure 1 As shown, a high-performance k-mer frequency counting method based on a clustering algorithm includes the following steps:
[0042] The CPU reads the genome format file and obtains the gene sequence reads of length L;
[0043] The CPU creates a CUDA stream that includes the following GPU kernel functions and the operations they perform:
[0044] S1: Split reads into L-K+1 k-mer sequences according to the preset subsequence length K;
[0045] S2: Take each base position in the k-mer sequence as a coordinate point, convert the multi-segment k-mer sequences obtained by splitting into corresponding coordinate arrays and add coordinate offsets;
[0046] S3: Use the clustering algorithm to calculate the centroid eigenvalues of each coordinate array, and return the centroid eigenvalues corresponding to all k-mer sequences and k-mer sequences to the CPU;
[0047] The CPU counts the frequency of occurrence of all k-mers according to the k-m...
Embodiment 2
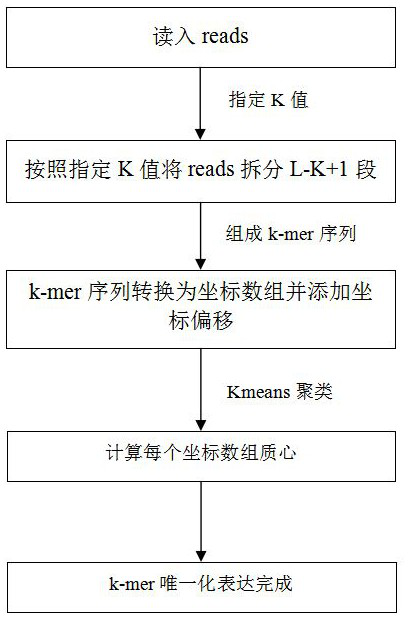
[0069] In this embodiment, based on the method of Embodiment 1, the Kmeans clustering analysis algorithm in the clustering algorithm is used to replace the mean-shift clustering algorithm to calculate the centroid. The specific centroid calculation process is as follows:
[0070] After converting the k-mer sequence into coordinate points, Kmeans clustering can be used to calculate the centroid of the set of coordinate points with the mean square error of the Euclidean distance between sample points (coordinate points) as the criterion function, which can be As the centroid feature value of the k-mer sequence, a k-mer sequence is uniquely expressed. At this point, all k-mer sequences of reads in the sequencing file can be converted into centroid expressions for subsequent statistics of the occurrence frequency of k-mers.
[0071] Among them, regarding the Kmeans cluster analysis algorithm, its related concepts include: (1) K value, that is, the number of clusters to be obtained...
Embodiment 3
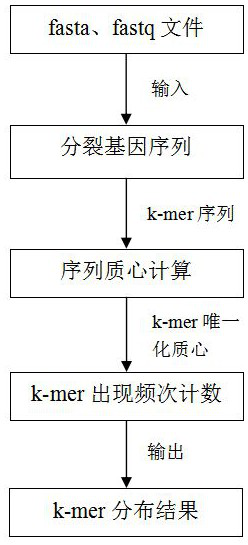
[0084] On the basis of the method provided in the first embodiment, the present invention further provides a high-performance k-mer frequency counting system based on a clustering algorithm. The system includes a k-mer preprocessing module, a coordinate transformation module, a Kmeans calculation module and a -mer frequency statistics module. in,
[0085] The K-mer preprocessing module is used to read the sequencing file from the disk through the CPU, transmit it to the GPU through the CUDA stream, and use the GPU to split the sequencing file into multiple k-mer sequences according to the preset subsequence length K.
[0086] The coordinate conversion module is used to take each base position in the k-mer sequence as a coordinate point, convert the multi-segment k-mer sequences obtained by splitting into corresponding coordinate arrays, and add coordinate offsets.
[0087] The Kmeans calculation module is used to calculate the centroid eigenvalues of each coordinate array u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com