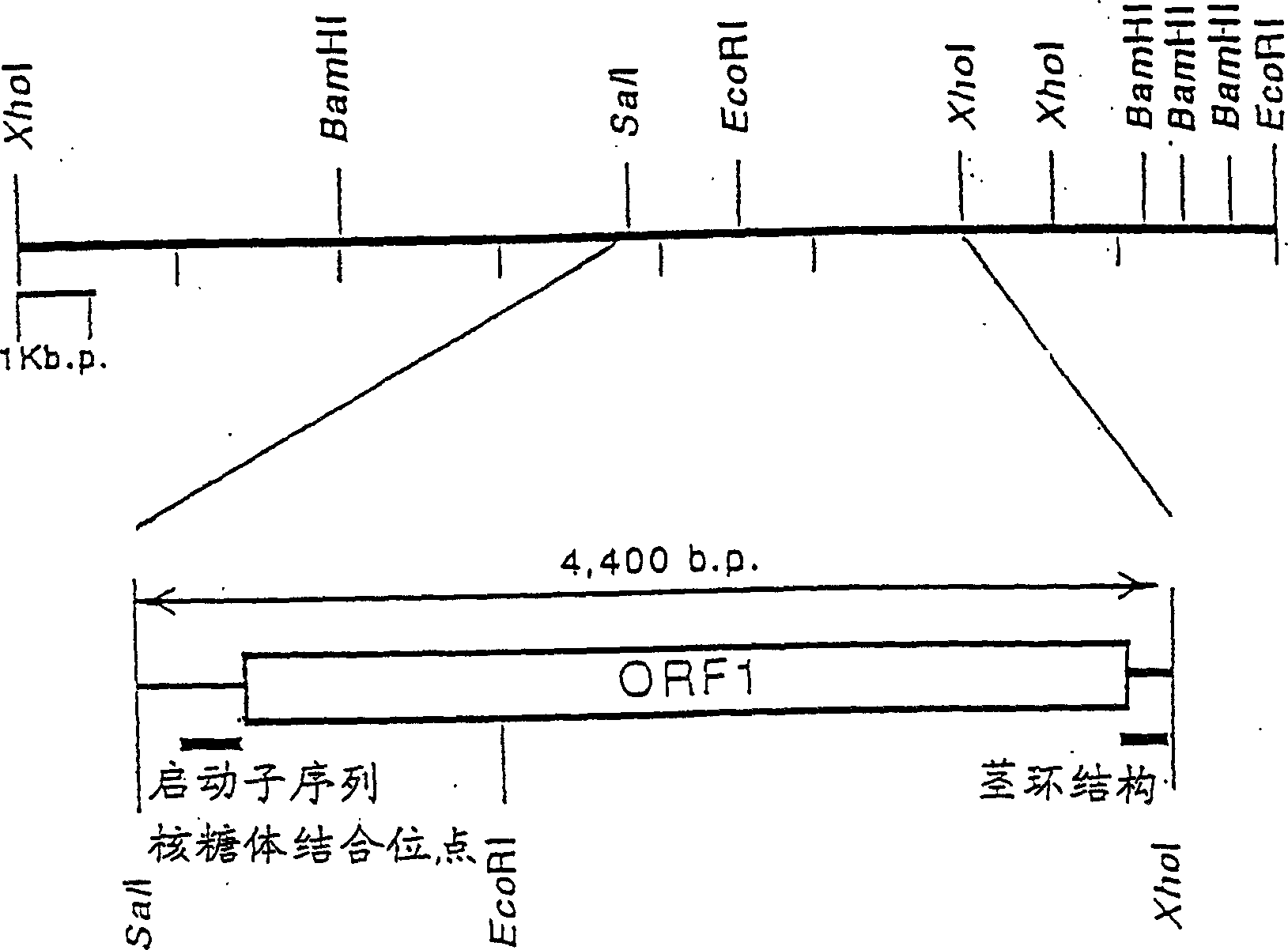
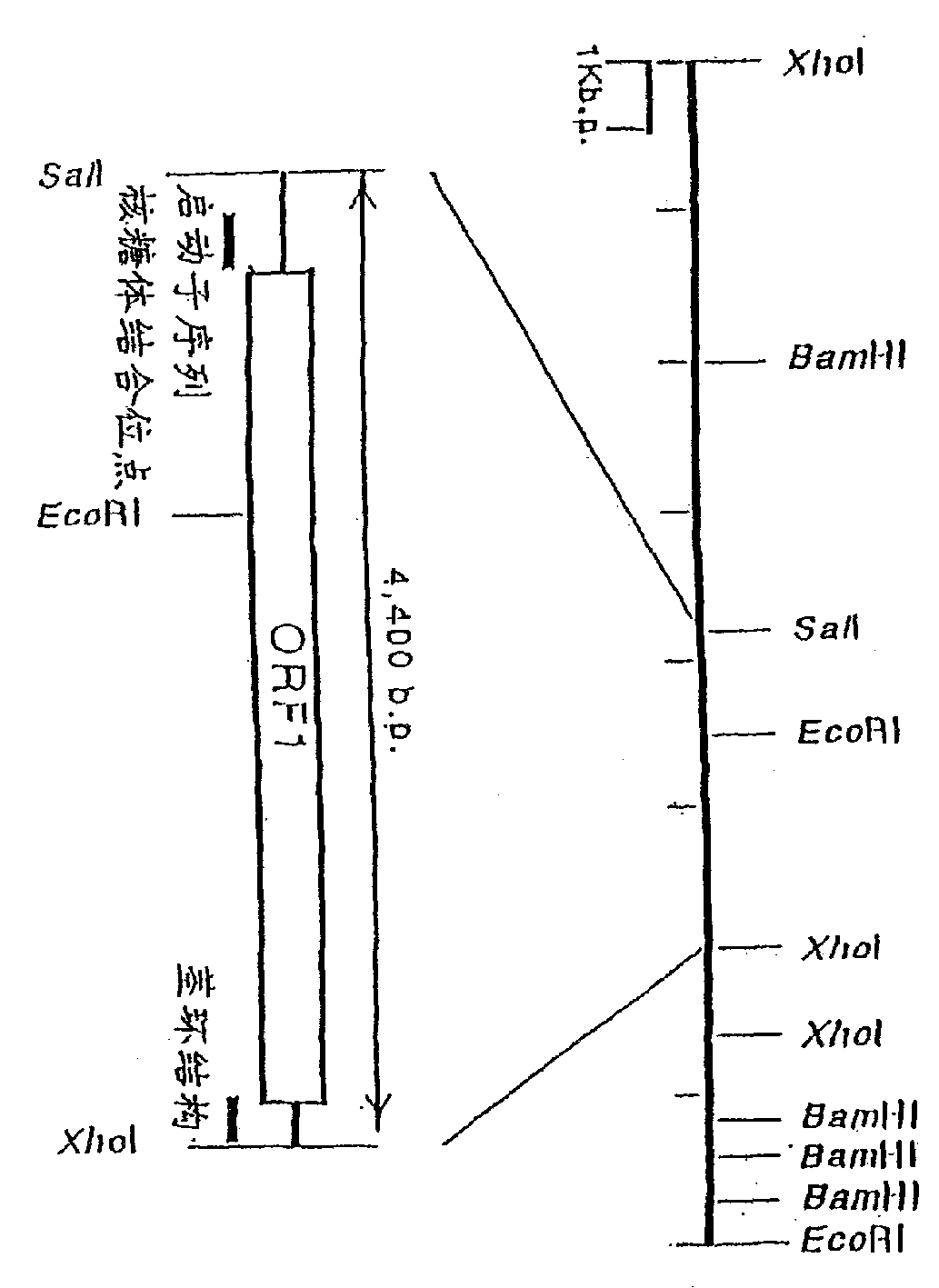
Alph-ketoglutaric acid dehydrase gene
A ketoglutarate dehydrogenase and gene technology is applied in the production field of alpha-ketoglutarate dehydrogenase gene and lysine, and can solve the problem of not finding coryneform bacteria mutants and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1: Isolation and structure determination of α-KGDH gene
[0100] (1) Preparation of probes
[0101] Selection of E. coli and B. subtilis α-KGDH E 1 There are regions of high homology between the subunit genes, and the phosphoramidite method was used to synthesize the oligonucleotides given by SEQ ID NOS.3 and 4 in the sequence listing with a DNA synthesizer (Model 394, manufactured by Applied Biosystems).
[0102] Oligonucleotides (0.25 μ mole) were used as primers, and chromosomal DNA (0.1 μ g) of Bacillus subtilis NA64 was prepared by a common method (this bacterial strain was obtained from Bacillus Genetic Stock Center (Ohio University, U.S.) as a template, Taq DNA polymerase (2.5 units) (manufactured by Jiubao Manufacturing Co., Ltd.) was added to 0.1 ml of 10 mM Tris-HCl buffer (pH 8.3) containing 200 μM each of dATP, dCTP, dGTP, and dTTp, 50 mM potassium chloride, 1.5 mM magnesium chloride and 0.0001% gelatin. Using the PCR method, each round includes...
Embodiment 2
[0117] Example 2: Increase of α-KGDH activity by expression of α-KGDH gene derived from Brevibacterium lactofermentum ATCC 13869
[0118] (1) Introducing the α-KGDH gene into Brevibacterium lactofermentum ATCC 13869 and AJ11060
[0119] The pHSGS-X plasmid DNA (1 μ g) that embodiment (1) obtains and restriction endonuclease Sal I and xho I (20 units each) was mixed with the buffer solution of (3) in Example (1), and reacted at 37° C. for 3 hours. On the other hand, plasmid pPK4 (refer to Japanese Laid-Open Patent No. 5-7491) DNA (1 μg) autonomously transcribed in Brevibacterium bacteria and Sal I (20 units) was mixed in the buffer solution of (3) in Example 1, and reacted at 37° C. for 3 hours. The two reaction solutions were subjected to phenol extraction and ethanol precipitation by conventional methods. Then, in order to prevent the DNA fragments derived from the plasmid vector from rejoining, the DNA fragments were dephosphorylated by bacterial alkaline phosphatase ...
reference example 1
[0128] Reference Example 1: Relationship between α-KGDH activity and L-glutamic acid production capacity
[0129] Brevibacterium lactofermentum AJ11060 / pPK4 and AJ11060 pPKS-X were cultured in the L-glutamic acid producing medium, and the L-glutamic acid produced and accumulated in the culture solution was measured. The cultivation was carried out by adding a surfactant as follows.
[0130] Contains 8% glucose, 0.1% potassium dihydrogen phosphate, 0.004% magnesium sulfate, 3% ammonium sulfate, 1.5% soybean hydrolyzate solution, 200μg / l thiamine hydrochloride, 300μg / l biotin, 25mg / l kanamycin and 5% CaCO 3 (Separately sterilized) production medium (pH 8.0, 20ml) is prepared and poured into a Sakaguchi flask with a volume of 500ml, and heat sterilized. Previously cultured on plates containing 1% polypeptone (manufactured by Nippon Pharmaceutical Co., Ltd.), 1% bacterial culture yeast extract (manufactured by Difco), 0.5% sodium chloride, 0.5% glucose and 25 mg / l kanamycin Bac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com