Preparation method of cholera toxin B sub unit
A technology of cholera toxin and subunit, which is applied in the field of producing CTB to prepare drugs, and can solve the problems of low expression, high production cost, cumbersome and multi-step post-treatment process, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
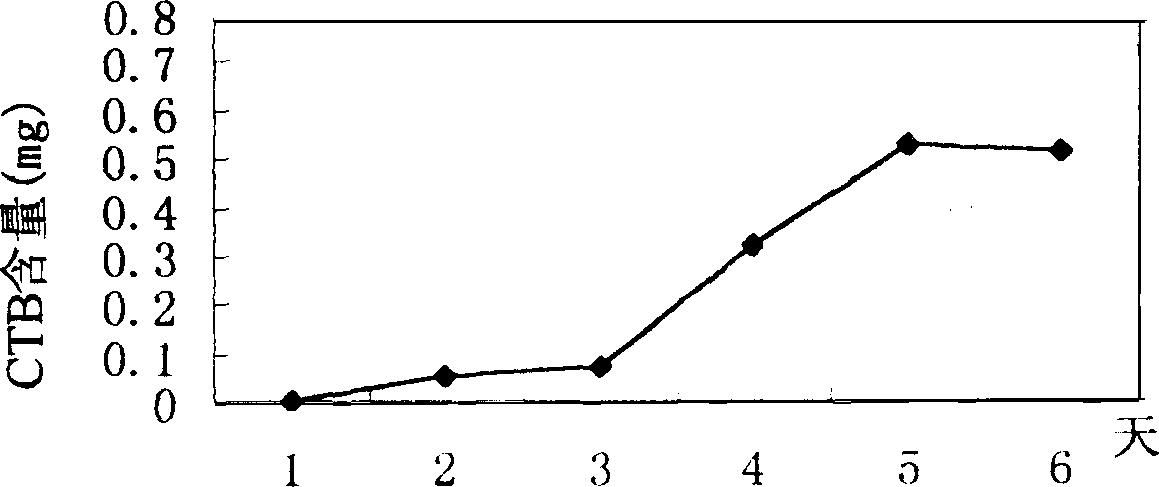
Image
Examples
Embodiment 1
[0024] Embodiment 1, the construction of CTB gene Escherichia coli expression plasmid
[0025] Primers were designed according to the published CTB gene sequence ((Nature, 1983, 306:551-557), the BamHI restriction site was introduced at the 5' end primer, and the targeting sequence and EcoR I site were introduced at the 3' end. The upstream primer For: 5'GGGGATCCATGATTAAATTAAAATTTGGT3', the downstream primer is: 5'GGGGAATTCTTATAGCTCATCTTTTCTCAGAATTTGCCATACTAATTGCGGCAA3', take Vibrio cholerae (Vibrio cholerae) 1ml, centrifuge at 12000rpm for 5 minutes, take the precipitate, add 5μl 20g / L proteinase K (GIBCOBRL company) and 10% SDS70μl, ℃, 1 hour, add 500 μl phenol, centrifuge at 12,000 rpm for 10 minutes, take the supernatant, add 500 μl of chloroform (Zhejiang Dier Pharmaceutical Co., Ltd.), centrifuge at 12,000 rpm for 10 minutes, take the supernatant, add 2 times the volume of ethanol, and mix well , centrifuged at 12000rpm for 10 minutes, discarded the supernatant, and adde...
Embodiment 2
[0026] The construction of the insect baculovirus transfer plasmid of embodiment 2, CTB gene
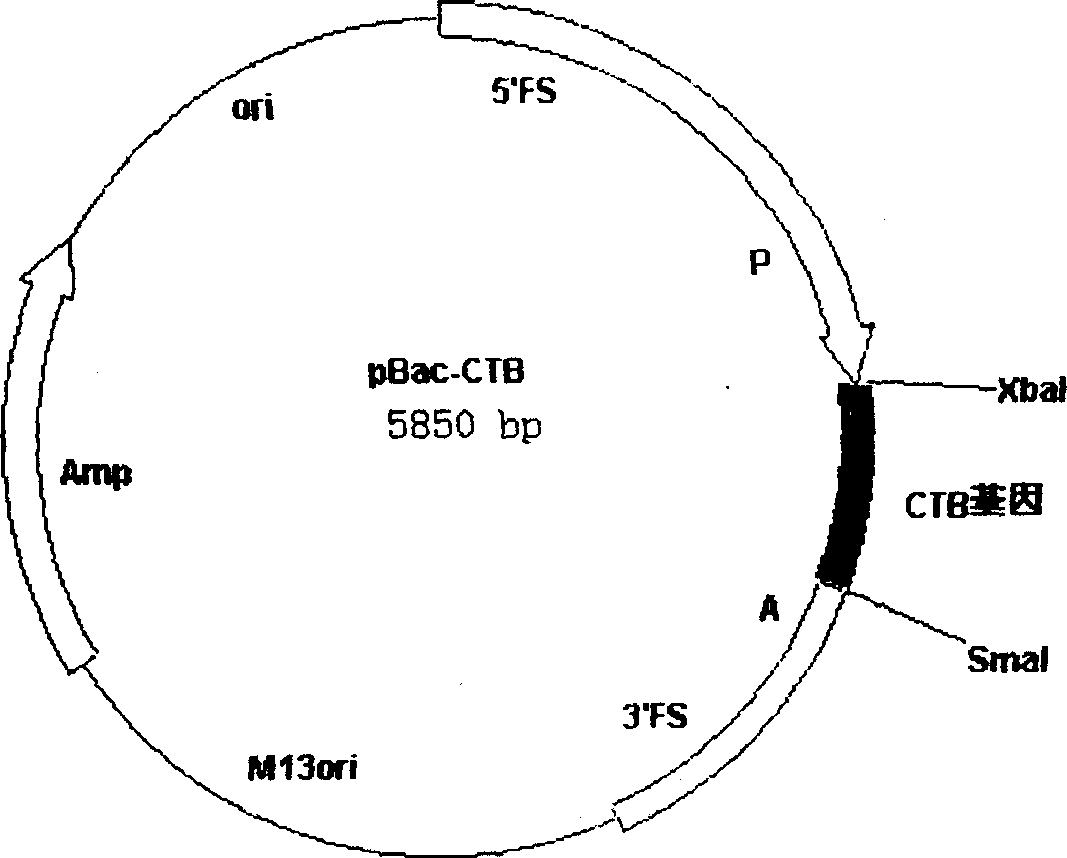
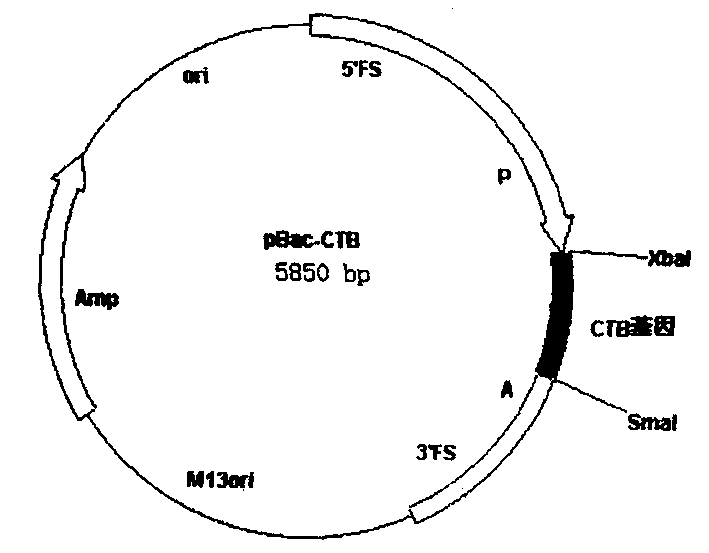
[0027] From the pUC-CTB plasmid digested with BamHI and EcoRI, the digested fragment was connected to pBacPAK8 (CLONTECH company) which was also digested with BamHI and EcoRI, and the bacmid transfer plasmid pBac-CTB containing the CTB gene was constructed ( figure 1 ), the gene was identified to be correct by restriction analysis.
Embodiment 3
[0028] Embodiment 3, the acquisition of the recombinant baculovirus of CTB gene
[0029] Take 5ul insect baculovirus transfer plasmid pBac-CTB containing CTB gene and 6ul wild silkworm nuclear polyhedrosis virus DNA for co-transfection. Take 6ul Lipofectin (GIBCOBRL company) and add 100ul serum-free TC-100 medium and mix well. The BmN cells previously cultured in a 35mm Dish were washed twice with serum-free TC-100 (GIBCOBRL company) medium, and the transfer plasmid and Lipofectin mixture was added dropwise, cultured at 27°C for 4-5 days, and the supernatant was collected for the second stage. One round of plaque screening. Take 5ul of the supernatant to infect the BmN cells in a 35mmDish, discard the supernatant after 1 hour and add an equal amount of mixed TC-100 medium and low melting point agarose. Pick plaques after 4-5 days, infect BmN cells for 3-4 days, save the supernatant, lyse the cells with NaOH for Southern hybridization, and take the supernatant of positive clo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com