Spiroplasma pathogenic microorganism PCR fast checking technique
A pathogenic microorganism and detection technology, which is applied in the field of PCR rapid detection technology for spiroplasma pathogenic microorganisms, can solve the problems of large sample volume, inability to quantify, unsuitable for live blood detection, etc., and achieve the effect of low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Embodiment 1: Detection of environmental sediment
[0016] In July 2004, a small amount of sediment samples were collected from a pond where crab shivering disease broke out in Quanjiao, Anhui Province, and brought back for testing. The specific operation is as follows:
[0017] a) Pretreatment: Dissolve about two medicine spoons of the mud sample to be tested in 50mL water for several hours, stir several times during the period to mix thoroughly → filter with quantitative filter paper to remove mud sample residue → put the filtrate in a 50mL centrifuge tube, 12000rpm , 4°C, centrifuge for 1 hour → carefully remove the supernatant, transfer the bottom liquid to a 1.5mL Eppendorf tube → 8000rpm 4°C, centrifuge for 1 hour → carefully remove the upper layer of liquid, leaving about 0.1mL in the tube;
[0018] b) Template extraction: (1) Add 150-200 μL of 5% Chelex-100 to the tube, shake vigorously for 10 seconds; (2) bathe in water at 56°C for 20 minutes, and mix several ...
Embodiment 2
[0021] Embodiment 2: detect the pathogenic bacteria of isolation culture
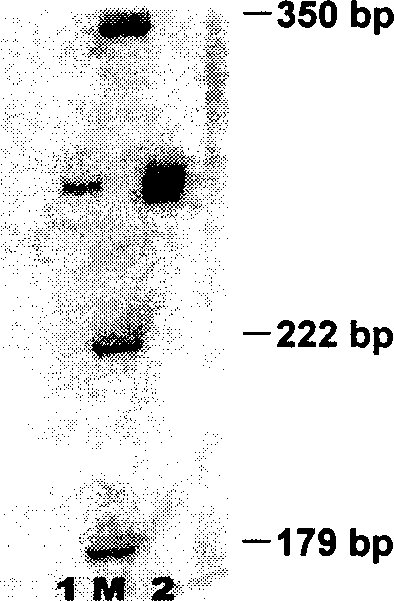
[0022] Take 1mL of the culture of spiroplasma pathogenic bacteria purely cultivated in the laboratory and dilute it to a spiroplasma concentration of 10 6 Individual / mL, 13000rpm, 4 ℃, centrifugal 30 minutes, remove supernatant; Template extraction, PCR reaction are the same as embodiment 1, 2% agarose electrophoresis as figure 2 Shown in track 6. figure 2 Middle M, Marker, from bottom to top are 100bp, 200bp, 300bp, 400bp, etc. The spiroplasma-specific band appears between 200bp and 300bp, which is 271bp.
[0023] According to the above method, the spiroplasma concentration was detected to be 10 4 pcs / mL, 10 2 samples / mL, the results are as follows figure 2 Tracks 7 and 8 are shown.
Embodiment 3
[0024] Example 3: Detection of host blood
[0025] During the high temperature in the summer of 2004, a few crabs suffering from shivering disease were collected.
[0026] a) Pretreatment: about 10 μL of blood was drawn from the living body, mixed in a small amount of PBS (pH=7.0), mixed evenly, and transferred into a 1.5 mL Eppendorf tube.
[0027] b) Template extraction: (1) Add 150-200 μL of 5% Chelex-100 to the tube containing the sample, and mix well with proteinase K (final concentration is 100 μg / mL); (2) 1.5-2 hours in a 56°C water bath, Mix several times during the period; (3) take out the vigorous shaking for 10 seconds, 99 ° C water bath for 8 minutes (strict control); (4) take out the vigorous shaking for 10 seconds, 13000 rpm, 4 ° C, centrifuge for 5 minutes; (5) transfer the supernatant ( DNA template) to another clean Eppendorf tube, stored at -20°C, and amplified as soon as possible.
[0028] c) PCR reaction is the same as in Example 1,
[0029] d) 2% agaros...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com