Yeast system capable of coexpressing CYP and CPR
A yeast expression system and co-expression technology, applied in the field of Saccharomyces cerevisiae new expression system, can solve the problems of unstable protein expression, low data repeatability, and system instability, and achieve the effects of easy construction, stable system, and reduced production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Construction and Identification of Example 1 Yeast Expression Plasmid
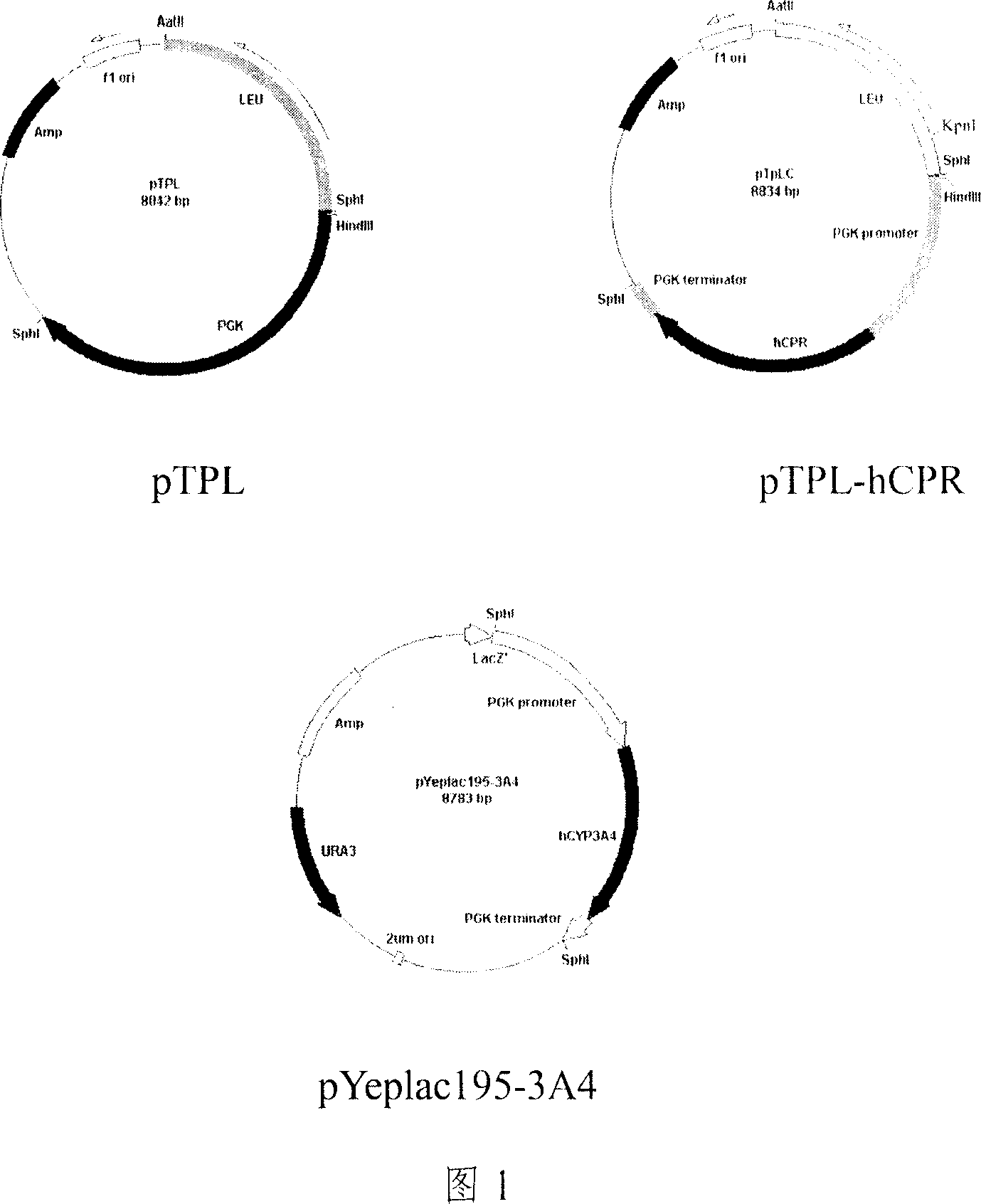
[0040] 1. Extract yeast chromosome and amplify to obtain PGK fragment. The obtained PGK amplified fragment was recovered and purified and directly ligated into the cloning vector pGEM-T easy vector, and the sequence of the obtained recombinant was confirmed to be correct. The vector pT-PGK uses Klenow enzyme to remove the NdeI site to obtain the pT-PGKn plasmid. LEU screening gene and pT-PGKn were digested by SacII and SphI, and then by T 4 DNA ligase ligation, transformation into Escherichia coli, the resulting recombinant is the pTPL plasmid (as shown in Figure 1). Using pTPL as a template, primers were designed for PCR amplification. Obtained pTPL' fragment, the length is about 5000bp (comprising the sequence of yeast promoter, terminator, yeast selection gene, Escherichia coli cloning vector).
[0041] 2. hCPR cDNA and hCYP1A2, hCYP2D6, hCYP2E1, hCYP2C9, hCYP3A4 cDNA were obtained by PCR am...
Embodiment 2
[0042] Example 2: Construction and identification of yeast expression system
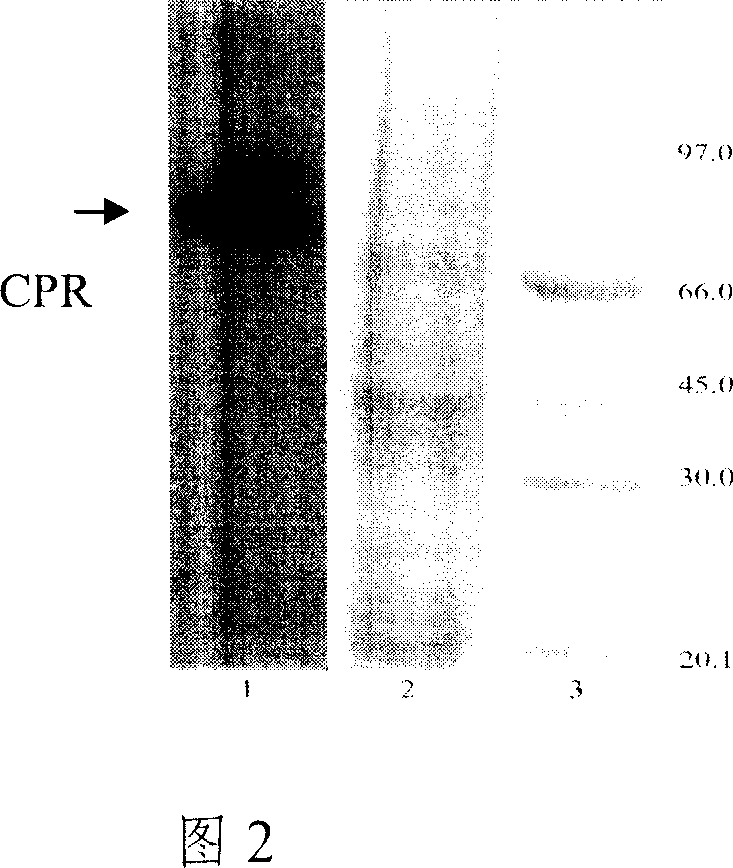
[0043] The plasmid pTPL-hCPR was linearized with SphI restriction endonuclease, and the yeast BWG1-7α competent cells were transformed. The recombinant was identified by leucine screening, and the host strain BWG-CPR was constructed. Yeast chromosomes were extracted and PCR amplified to identify recombinants. At the same time, yeast microsomes were prepared, microsomes were electrophoresed by SDS-PAGE, and CPR expression bands were analyzed by western blot (as shown in Figure 2).
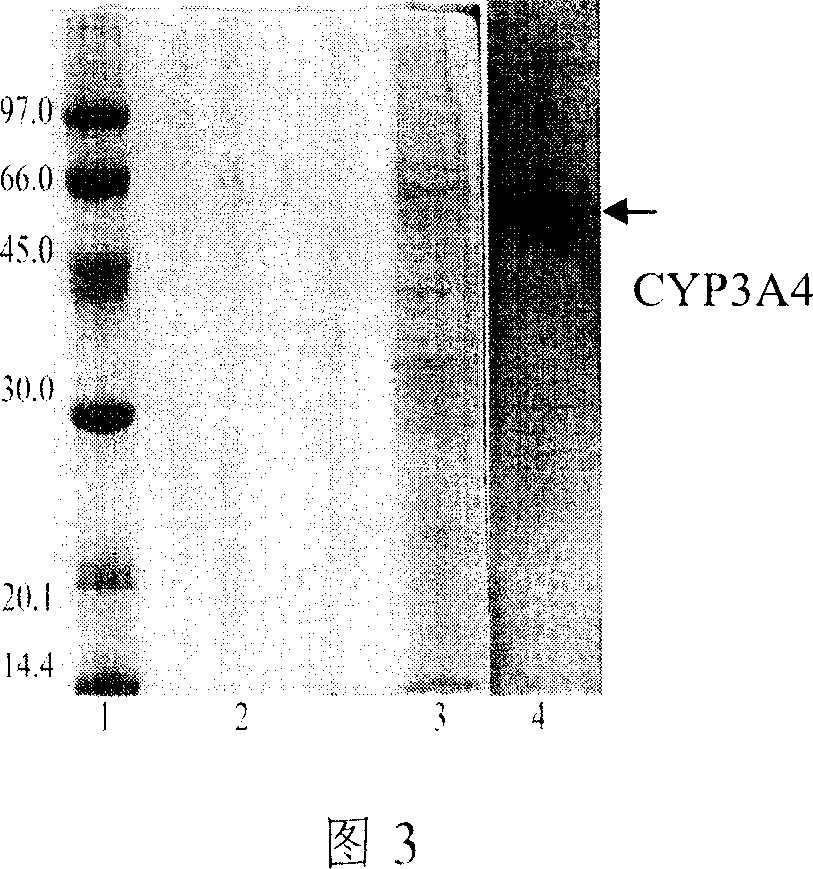
[0044] The plasmid pYeplac195-CYP was directly transformed into BWG-CPR competent cells, and the recombinant was screened and identified with uracil, and the strain BWG-CPR / CYP was constructed. Pick transformed colonies and inoculate them in liquid YPD medium, and culture them at 30°C until OD 600 =0.6-0.8, yeast chromosomes were extracted and PCR amplified to identify recombinants. At the same time, yeast microsomes w...
Embodiment 3
[0045] Example 3: Expression of protein in yeast and analysis of system activity and stability
[0046] 1. Preparation of yeast microsomes (yeast microsome, YM)
[0047] Fermentation medium 1L, medium components: 10g of yeast extract, 20g of peptone, 30g of glucose, distilled in 1000ml of distilled water.
[0048] When the cells start to shake, add cell δ-ALA to make the final concentration 0.5mmol / L. Shake at 30°C for 6 hours, cool down to 25°C, shake for 30 minutes, then rapidly raise the temperature to 30°C until the cells grow to OD 600 =1.0-1.5, centrifuge the bacterial liquid, collect the bacterial cells, and prepare yeast microsomes.
[0049] 2. Activity determination and stability analysis of yeast system
[0050] The Bradford method was used to detect the protein content of yeast microsomal, and to detect the CPR activity (Table 4) and CYP content in the microsome (as shown in Figure 4, CYP3A4-CO reduction absorption peak). The expression levels of hCYP1A2, hCYP2D...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com