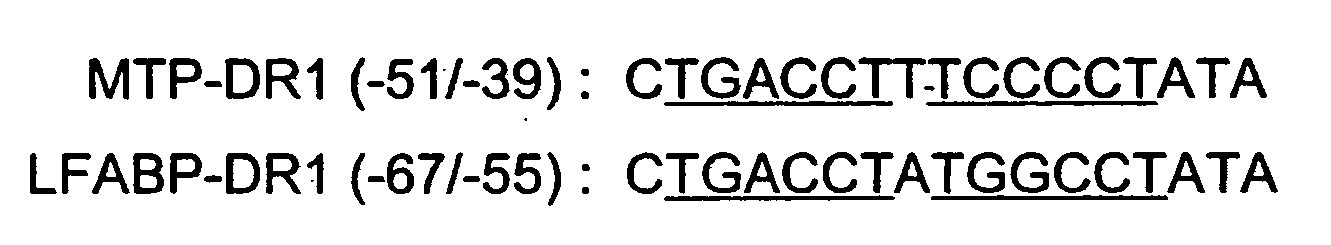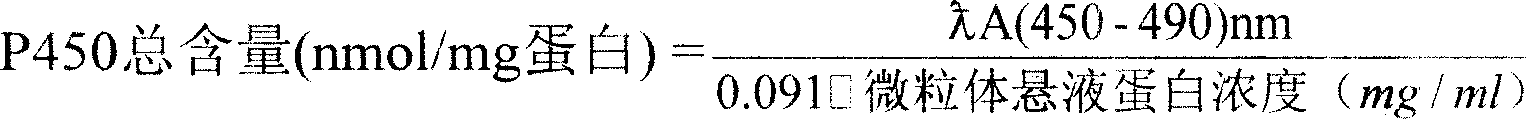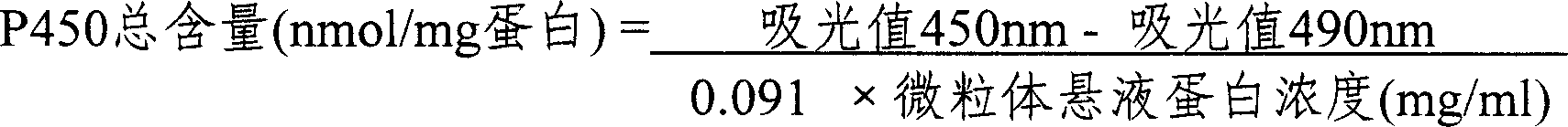Patents
Literature
96 results about "Microsome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
In cell biology, microsomes are heterogenous vesicle-like artifacts (~20-200 nm diameter) re-formed from pieces of the endoplasmic reticulum (ER) when eukaryotic cells are broken-up in the laboratory; microsomes are not present in healthy, living cells.
Pharmaceutical combinations for lipid management and in the treatment of atherosclerosis and hepatic steatosis
A pharmaceutical combination comprising an effective amount of at least one cholesterol absorption inhibitor and at least one microsomal triglyceride transfer protein inhibitor (MTP).
Owner:SCHERING CORP
Air purifying ball and preparation method thereof
The invention provides an air purifying ball. Various mineral elements are prepared into spherical particles which have multiple pores with the porosity of more than 65 percent and contain rich active genes by composite processing. Different components of the spherical particles can be used for natural absorption chemical combination, decomposition and diffusion in any space. The air purifying ball is suitable for air purification in any space and purified air in a forced ventilation filter. The invention can also extract effective components-air active microsomes which can purify air and improve air quality from about 60 plants by using the spherical particles as carriers and using processes, such as distillation, backflow, press squeezing, supercritical fluid extraction, and the like, select and form the combination of the air active microsomes according to different odor classes of an odor source to form active liquid-an air solvent by composite processing so as to carry or infiltrate the spherical particles so that the air purifying ball is formed.
Owner:傅朝春
Genes for microsomal delta-12 fatty acid desaturases and related enzymes from plants
InactiveUS6919466B2Control levelControl natureAnalysis using chemical indicatorsOther foreign material introduction processesLipid compositionFatty Acid Desaturases
The preparation and use of nucleic acid fragments encoding fatty acid desaturase enzymes are described. The invention permits alteration of plant lipid composition. Chimeric genes incorporating such nucleic acid fragments with suitable regulatory sequences may be used to create transgenic plants with altered levels of unsaturated fatty acids.
Owner:EI DU PONT DE NEMOURS & CO
Combination therapy for treating obesity or maintaining weight loss
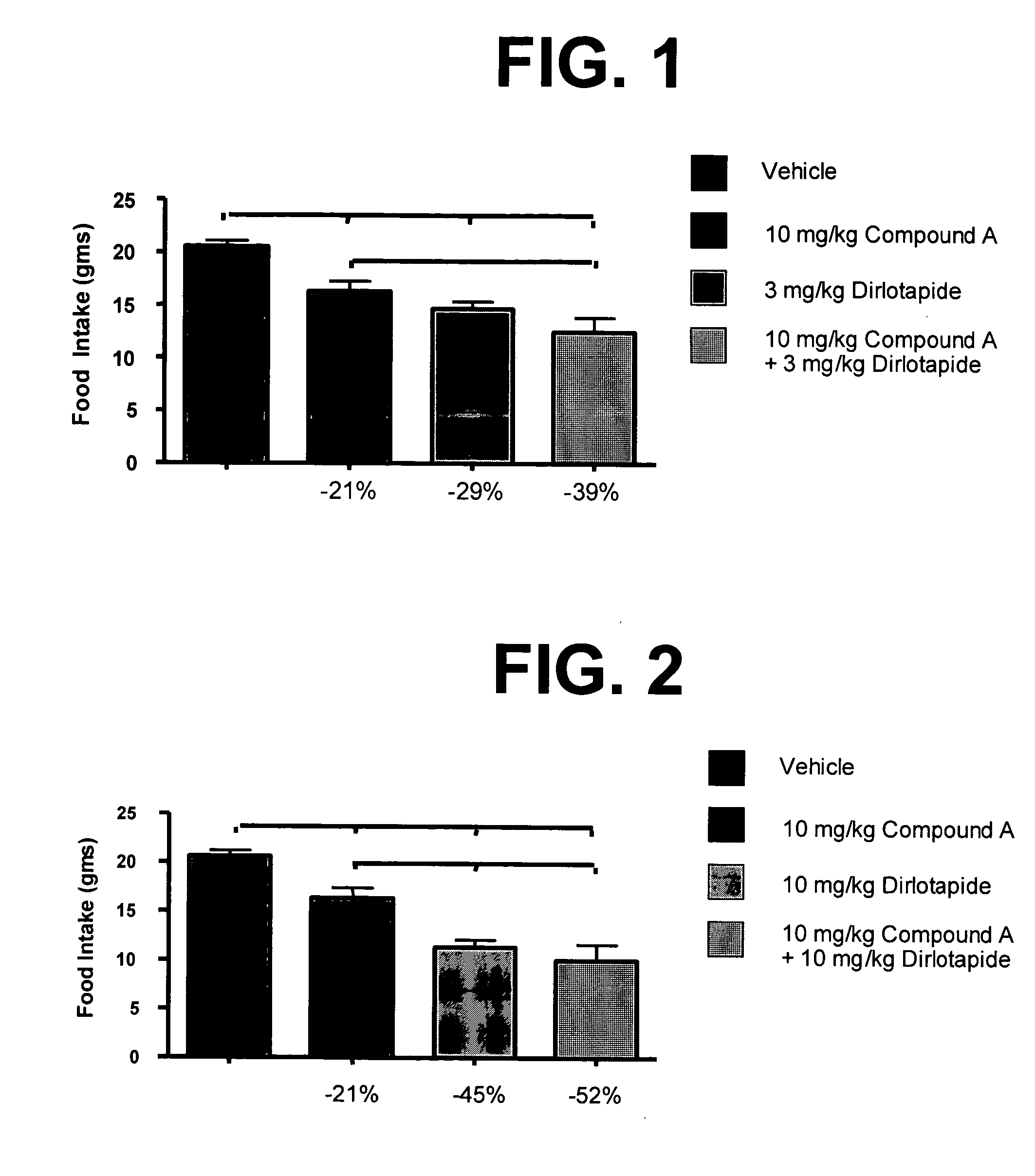
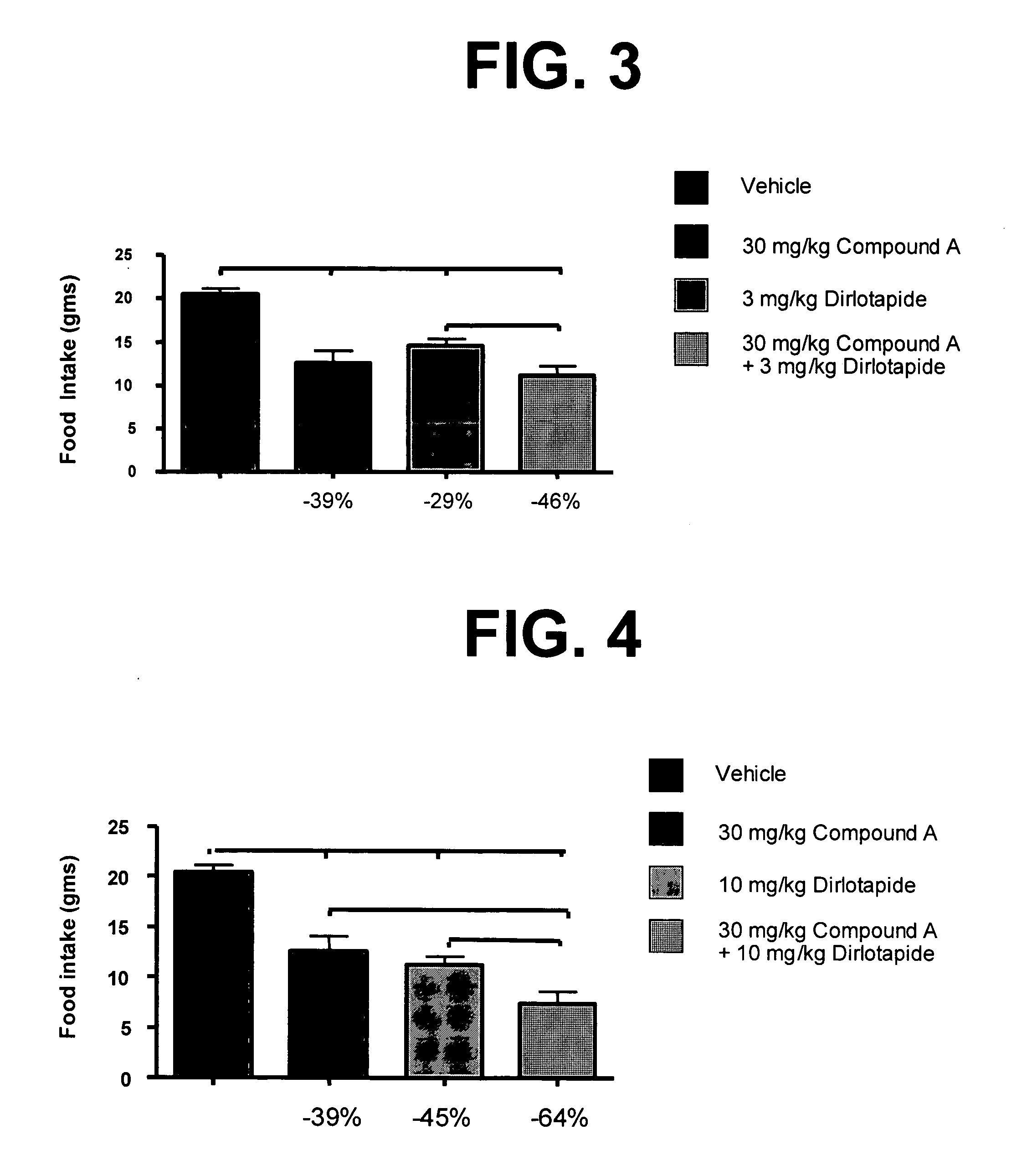
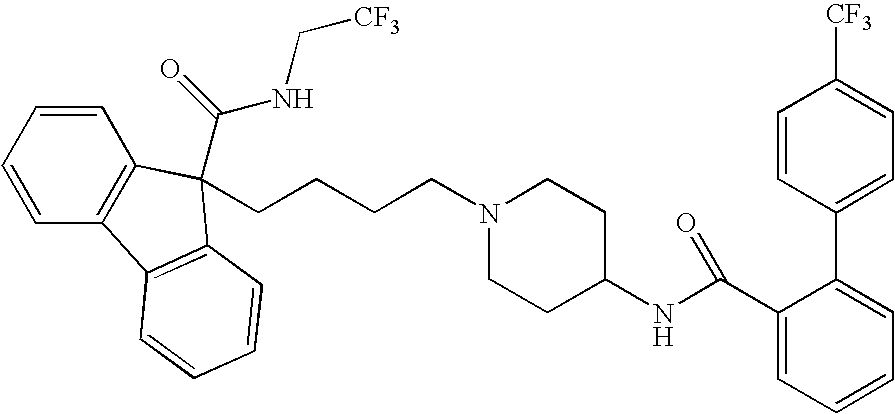
Combination therapies for treating obesity or related eating disorders and / or reducing food consumption are described herein which comprises administering a therapeutically effective amount of a cannabinoid-1 (CB-1) receptor antagonist and an intestinal-acting microsomal triglyceride transfer protein inhibitor (MTPi) to an animal in need of such treatment. The CB-1 receptor antagonist and intestinal-acting MTPi may be administered separately or together.
Owner:SWICK ANDREW G +1
Methods for treating disorders associated with hyperlipidemia in a mammal
InactiveUS20070093468A1Reduce concentrationExcessive amountBiocideSenses disorderCo administrationSide effect
Owner:AEGERION PHARM INC
Methods for treating disorders associated with hyperlipidemia in a mammal
The invention is directed to methods for treating hyperlipidemia in a mammal. The methods involve combination therapies using a microsomal triglyceride transfer protein (MTP) inhibitor (for example, BMS-201038 and implitapide) and a HMG-CoA reductase inhibitor (for example simvastatin or atorvastatin). Co-administration of the MTP inhibitor with the HMG-CoA reductase inhibitor produces a therapeutic benefit, for example, a reduction in the concentration of cholesterol and / or triglycerides in the blood stream, but with fewer or reduced side effects than when higher dosages of the MTP inhibitor are used during monotherapy to provide the same or similar therapeutic benefit.
Owner:AEGERION PHARM INC
Compositions and methods for ameliorating hyperlipidemia
InactiveUS20080241869A1Decreased plasma lipidReduce hyperlipidemiaOrganic active ingredientsMetabolism disorderSteatosisSecondary hyperlipidemia
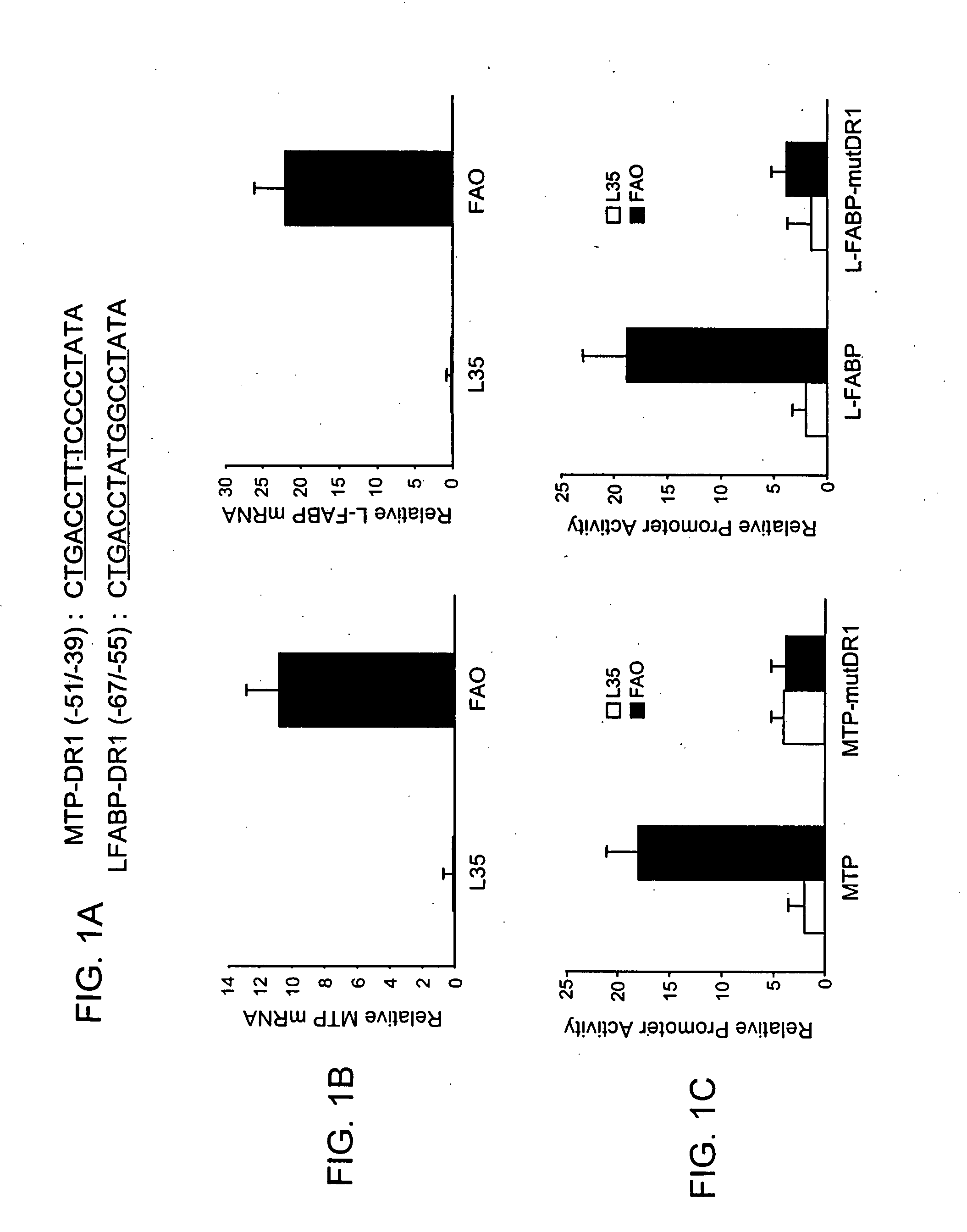
The invention provides compositions and methods for treating hyperlipidemia by administering a Microsomal Triglyceride Transfer Protein (MTP) inhibitor in combination with a Liver Fatty Acid-Binding Protein (L-FABP) inhibitor, methods of preventing the development of hepatic steatosis, methods of identifying an agent useful for treating hyperlipidemia, and methods of screening for inhibitors of MTP and L-FABP activity. Also provided are pharmaceutical compositions comprising a MTP inhibitor and a L-FABP inhibitor.
Owner:SAN DIEGO STATE UNIV RES FOUND
Compositions for Lowering Serum Cholesterol and/or Triglycerides
InactiveUS20080253985A1Reduce incidenceReduce severityBiocideSenses disorderHMG-CoA reductaseDepressant
The invention provides methods and compositions for treating hyperlipidemia and disorders associated with hyperlipidemia in a mammal. Compositions useful in the practice of the invention include a microsomal triglyceride transport protein inhibitor (“MTPI”) and at least two other cholesterol lowering drugs selected from the group consisting of a cholesterol absorption inhibitor (“CAI”), a HMG-CoA reductase inhibitor, a bile acid sequestrant, a fibric acid derivative, niacin, and squalene synthetase inhibitor.
Owner:AEGERION PHARM INC
Detection method for simultaneously determining metabolic products of seven CYP450 enzyme probe substrates in human liver microsomes
InactiveCN104849371AThe pretreatment method is simpleSuitable for routine testingComponent separationMetaboliteHepatica
The invention relates to a detection method for simultaneously determining the metabolic products of seven CYP450 enzyme probe substrate in human liver microsomes, and belongs to the technical field of biological detection. The detection method comprises the following steps: performing incubation on specificity probe substrates of CYP450 enzyme and the human liver microsomes for different periods of reaction time to generate corresponding metabolic products, using Zaltoprofen as an interior label, adopting a high performance liquid chromatography-tandem mass spectrometry after protein precipitation pretreatment, and thus simultaneously detecting the concentrations of the metabolic products of seven probe substrates. The method disclosed by the invention is high in specificity, high in sensitivity and simple and convenient in operation, and has already successfully been applied to the research of baicalin on CYP450 enzyme inhibiting action of the human liver microsomes.
Owner:WUXI PEOPLES HOSPITAL
Antisense modulation of microsomal triglyceride transfer protein expression
InactiveUS20050181376A1Sugar derivativesPeptide/protein ingredientsTriglycerideMicrosomal triglyceride transfer protein
Antisense compounds, compositions and methods are provided for modulating the expression of microsomal triglyceride transfer protein. The compositions comprise antisense compounds, particularly antisense oligonucleotides, targeted to nucleic acids encoding microsomal triglyceride transfer protein. Methods of using these compounds for modulation of microsomal triglyceride transfer protein expression and for treatment of diseases associated with expression of microsomal triglyceride transfer protein are provided.
Owner:IONIS PHARMA INC
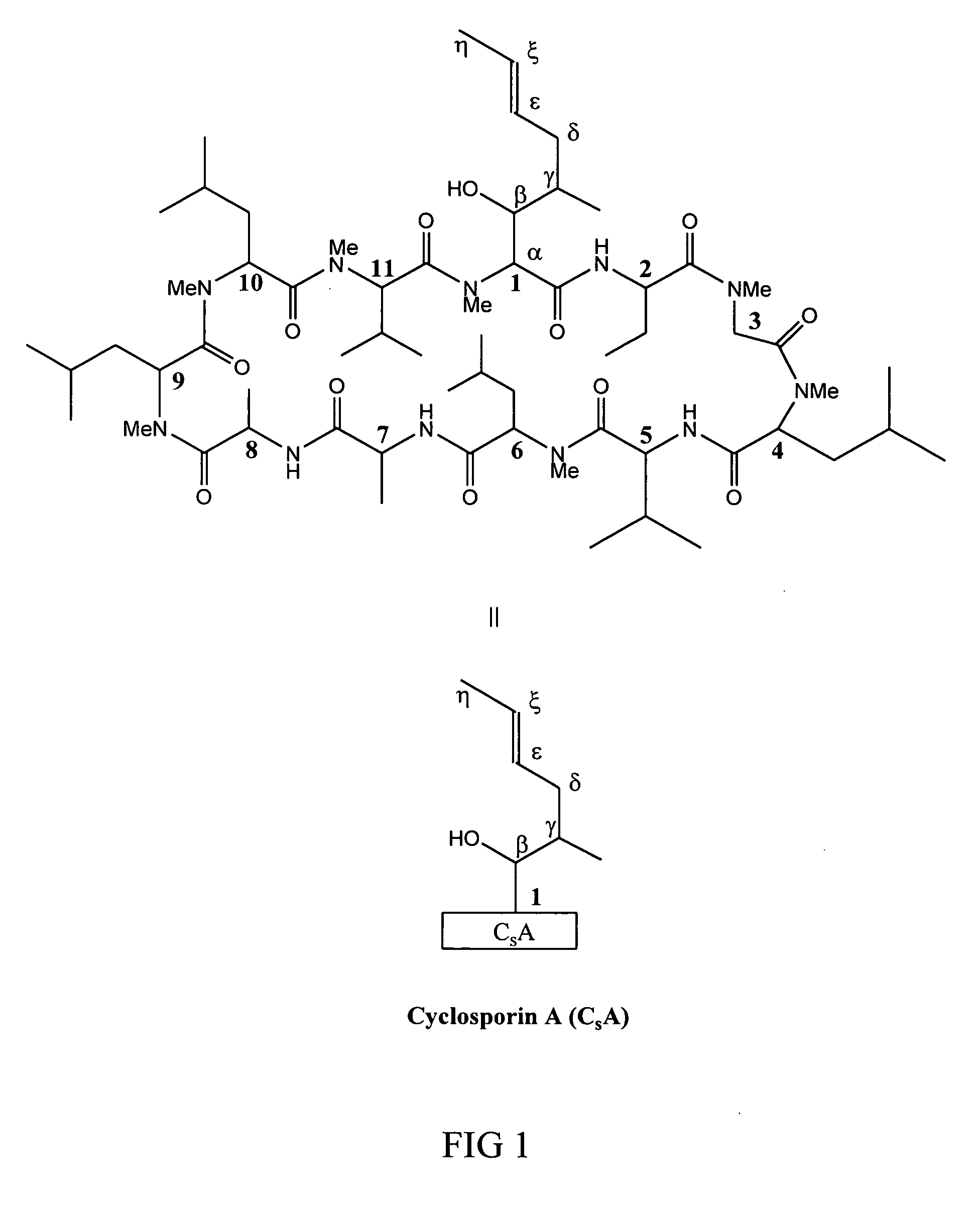
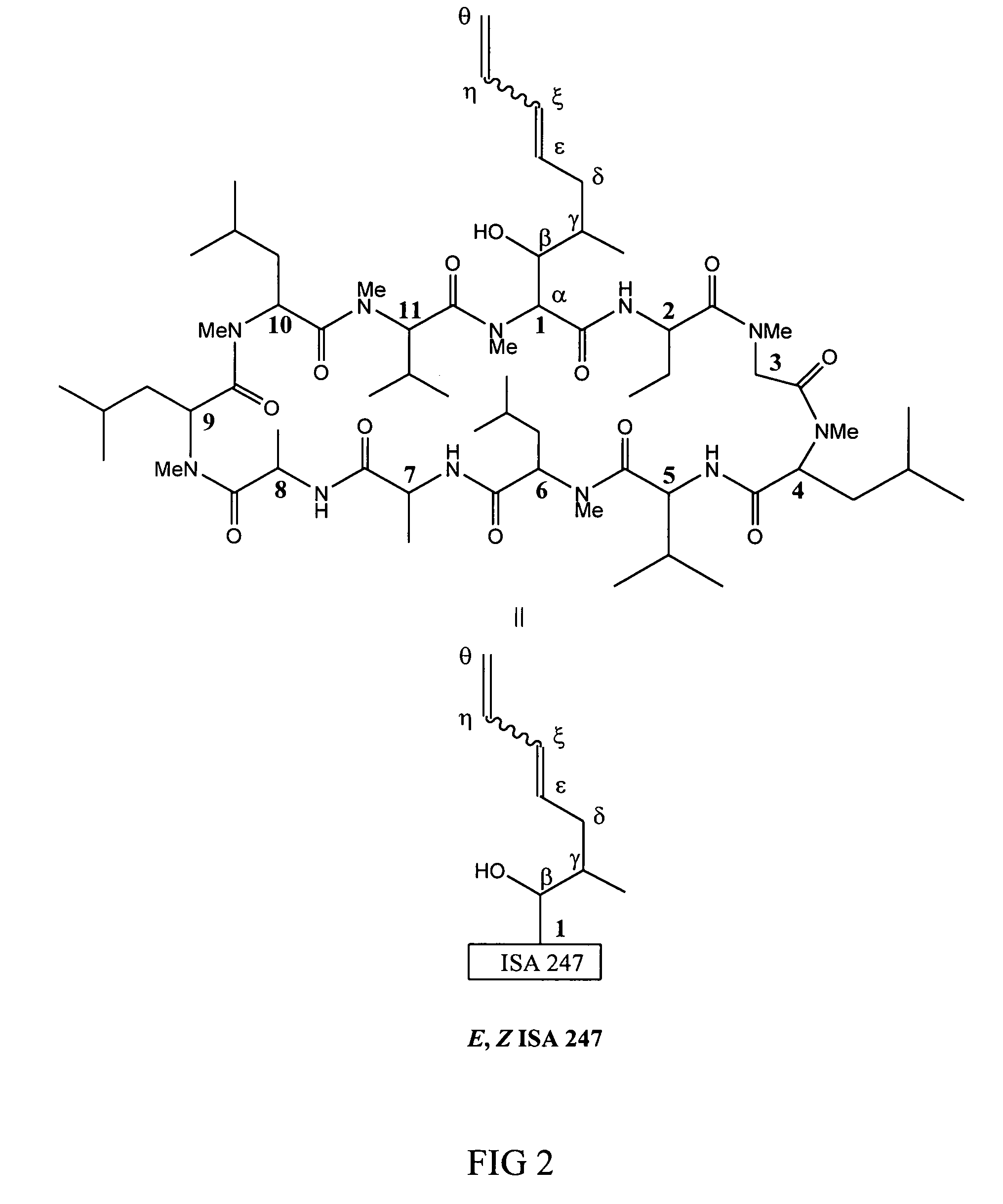
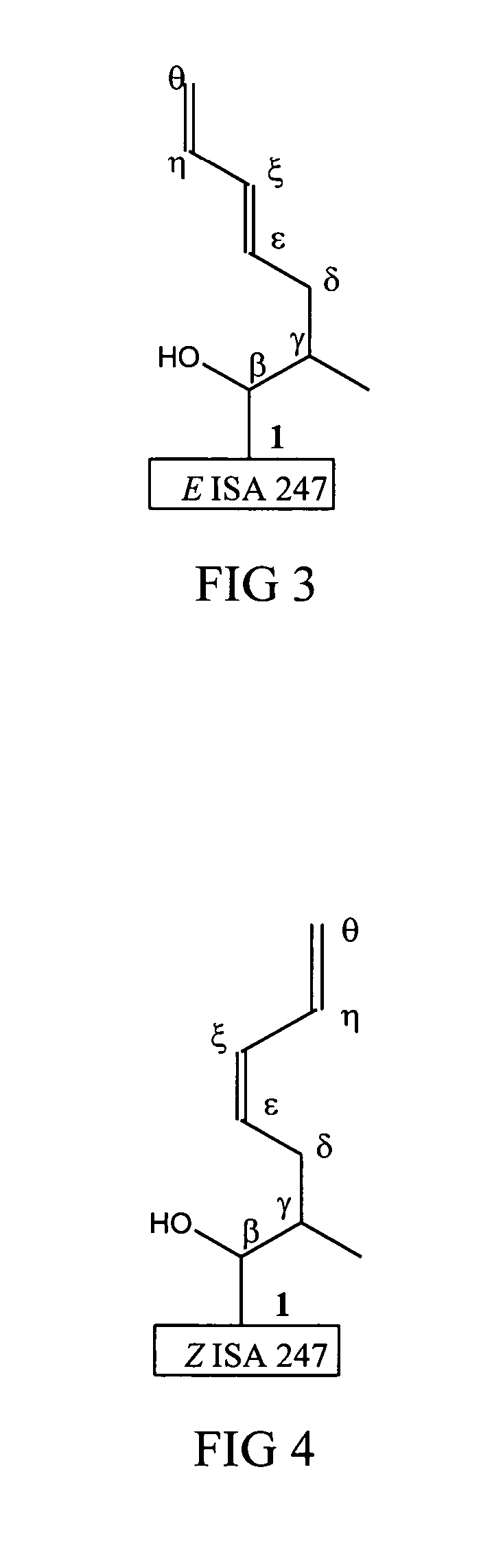
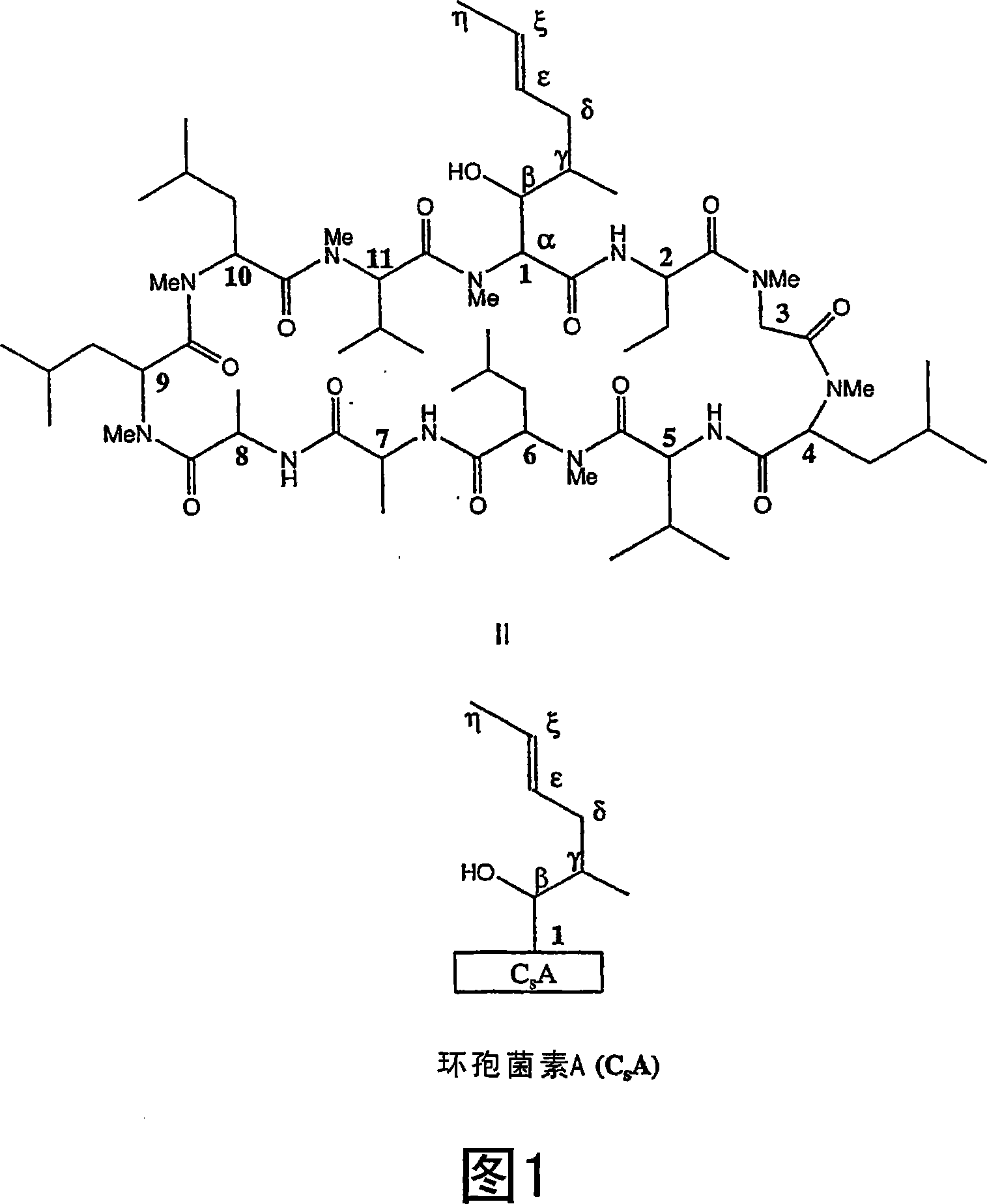
Metabolites of cyclosporin analogs
InactiveUS20060223743A1Useful immunosuppressive activityUseful for developmentCyclic peptide ingredientsImmunoglobulinsPhosphorylationCyclosporins
Isolated metabolites of the cyclosporine analog ISA247 are disclosed, including in vitro methods for their preparation. The metabolites comprise a chemical modification of ISA247, wherein the modification is at least one reaction selected from the group consisting of hydroxylation, N-demethylation, diol formation, epoxide formation, and intramolecular cyclization phosphorylation, sulfation, glucuronide formation and glycosylation. Methods of preparation include semi-synthetic methods, wherein metabolites of ISA247 are produced from the microsomal extracts of animal liver cells, or from cultures using microorganisms, and completely synthetic methods, such as chemically modifying the parent compound or isolated metabolites using organic synthetic methods.
Owner:ISOTECHNIKA INC
Genes for microsomal delta-12 fatty acid desaturases and hydroxylases from plants
InactiveUS7105721B2Analysis using chemical indicatorsOther foreign material introduction processesLipidomeHydroxylase gene
The preparation and use of nucleic acid fragments encoding fatty acid desaturase enzymes are described. The invention permits alteration of plant lipid composition. Chimeric genes incorporating such nucleic acid fragments with suitable regulatory sequences may be used to create transgenic plants with altered levels of unsaturated fatty acids.
Owner:EI DU PONT DE NEMOURS & CO
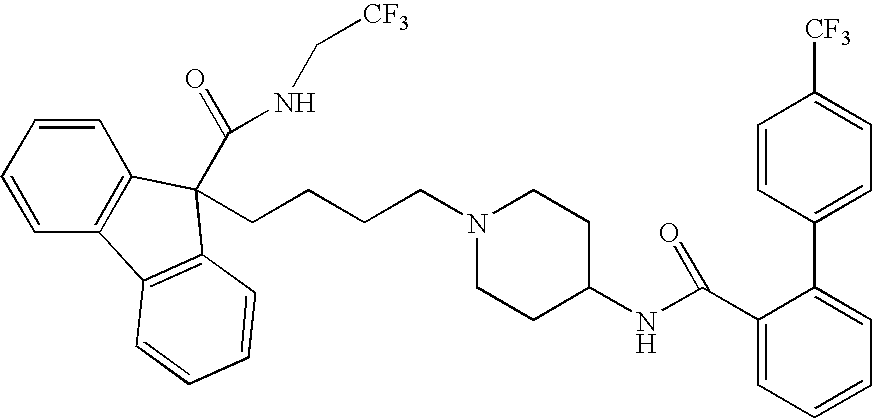
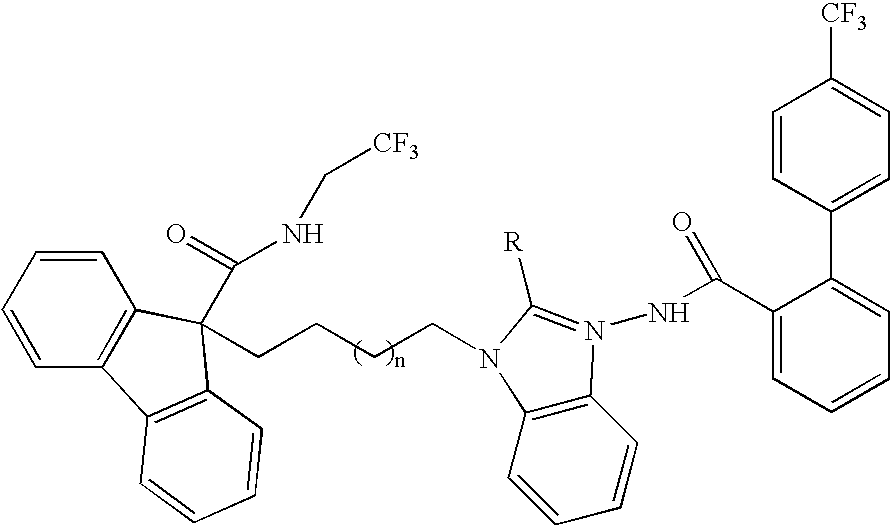
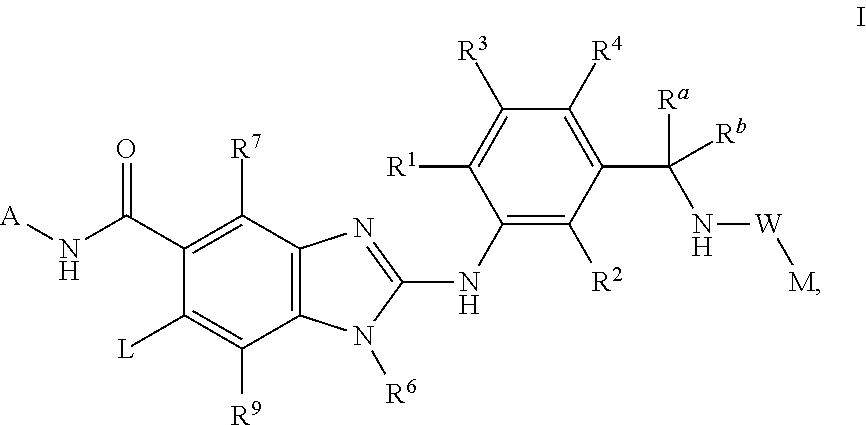
Novel compounds
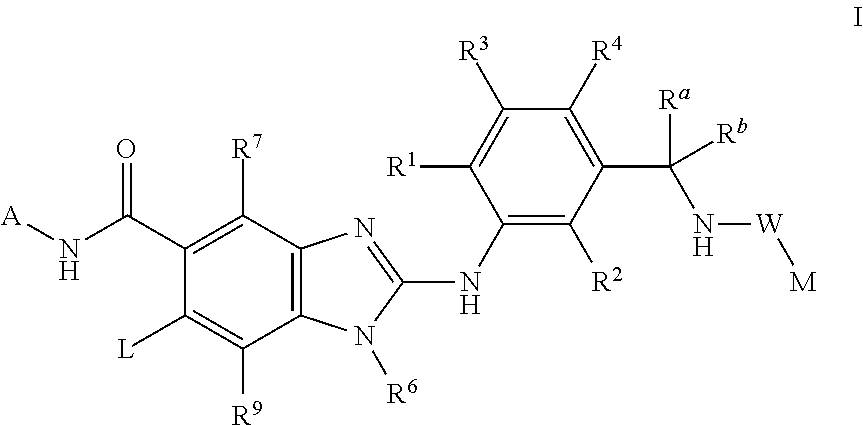
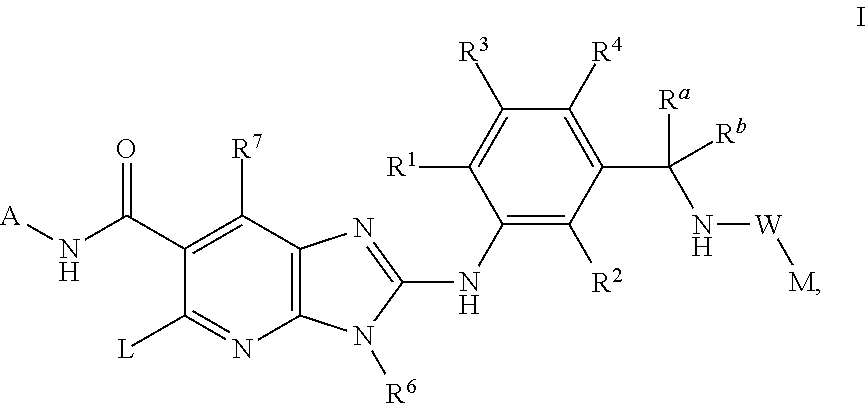
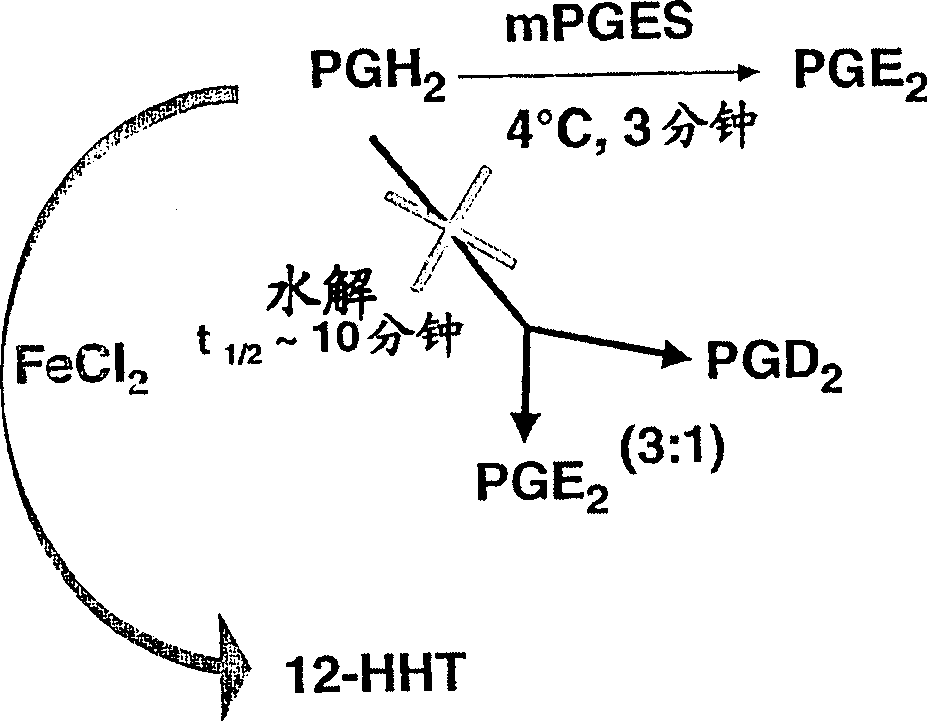
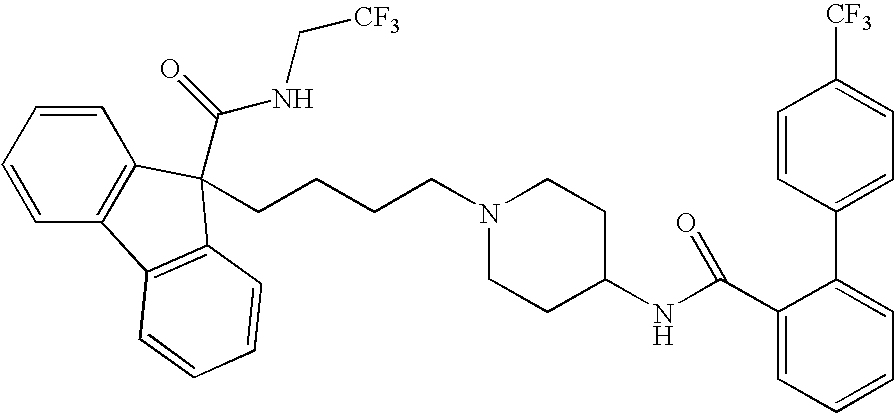
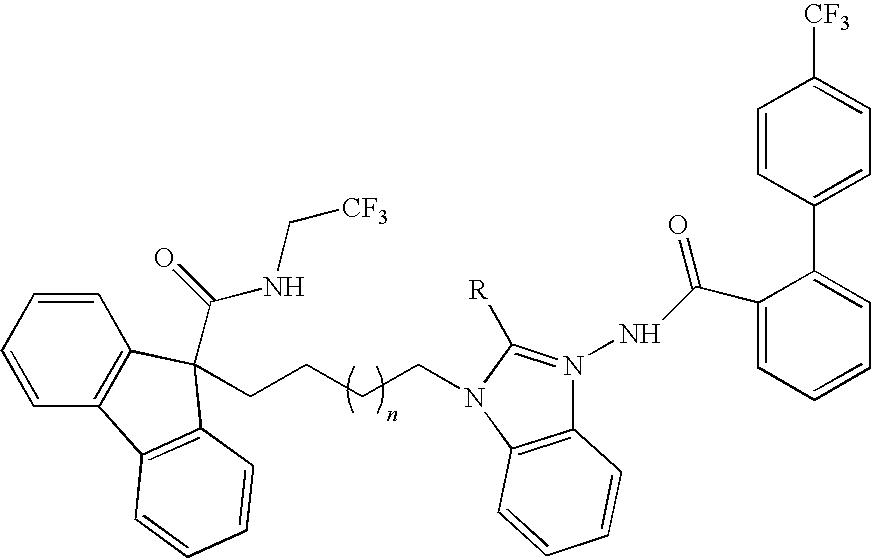
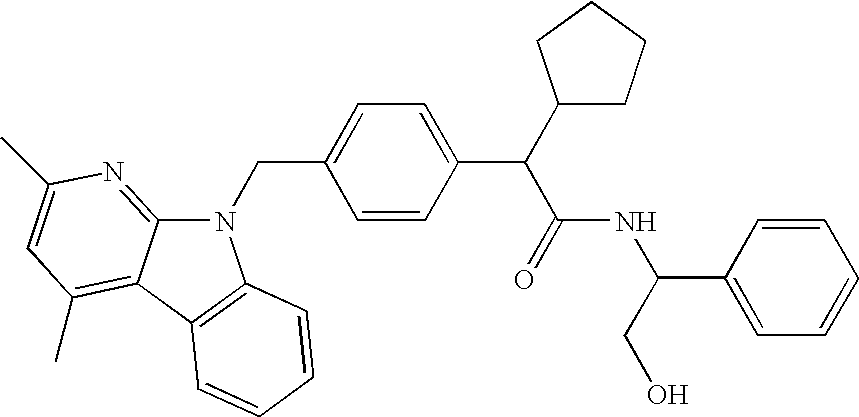
This invention relates to compounds of formula Itheir use as inhibitors of the microsomal prostaglandin E2 synthase-1 (mPGES-1), pharmaceutical compositions containing them, and their use as medicaments for the treatment and / or prevention of inflammatory diseases and associated conditions. A, L, M, W, R1, R2, R3, R4, R6, R7, R9, Ra, Rb have meanings given in the description.
Owner:BOEHRINGER INGELHEIM INT GMBH
Method and detector for detecting tumor microsomes by using laser tweezers and micro fluidics
The invention relates to a method and equipment for detecting tumor microsomes, and especially relates to and discloses a method and a detector for detecting tumor microsomes by using laser tweezers and micro fluidics. The detection method comprises the steps that: laser tweezers of different polarizations are generated; dual-beam double laser tweezers are used for focusing, and particles combined with tumor microsome antibodies in a channel of a micro fluidic chip are captured by the tweezers; a sample to be detected is mixed with the particles combined with tumour microsome antibodies; changes of physical parameters of the particles combined with tumor microsome antibodies before and after being mixed with the sample are collected by the detector, and qualification and accurate quantitation of the sample requiring detection can be carried out according to the parameters. The detector comprises a laser system, a nanometer positioning system, a camera system, optical parts, mechanical parts, a data collecting and transmitting system, and automatic controlling and image processing application software. According to the invention, single tumor microsome particle can be detected, and the method and the equipment are characterized in high specificity, simple operation, short detection period, high sensitivity, and good repeatability. Also, with the method and the equipment, blood samples can be re-detected.
Owner:嘉兴文斌生物技术有限公司 +1
Chiral separation of benzoporphyrin derivative mono-and di-acids by laser-induced fluorescence capillary electrophoresis
InactiveUS6331235B1Easy to detectEasy to separateElectrolysis componentsVolume/mass flow measurementCapillary electrophoresisRelative standard deviation
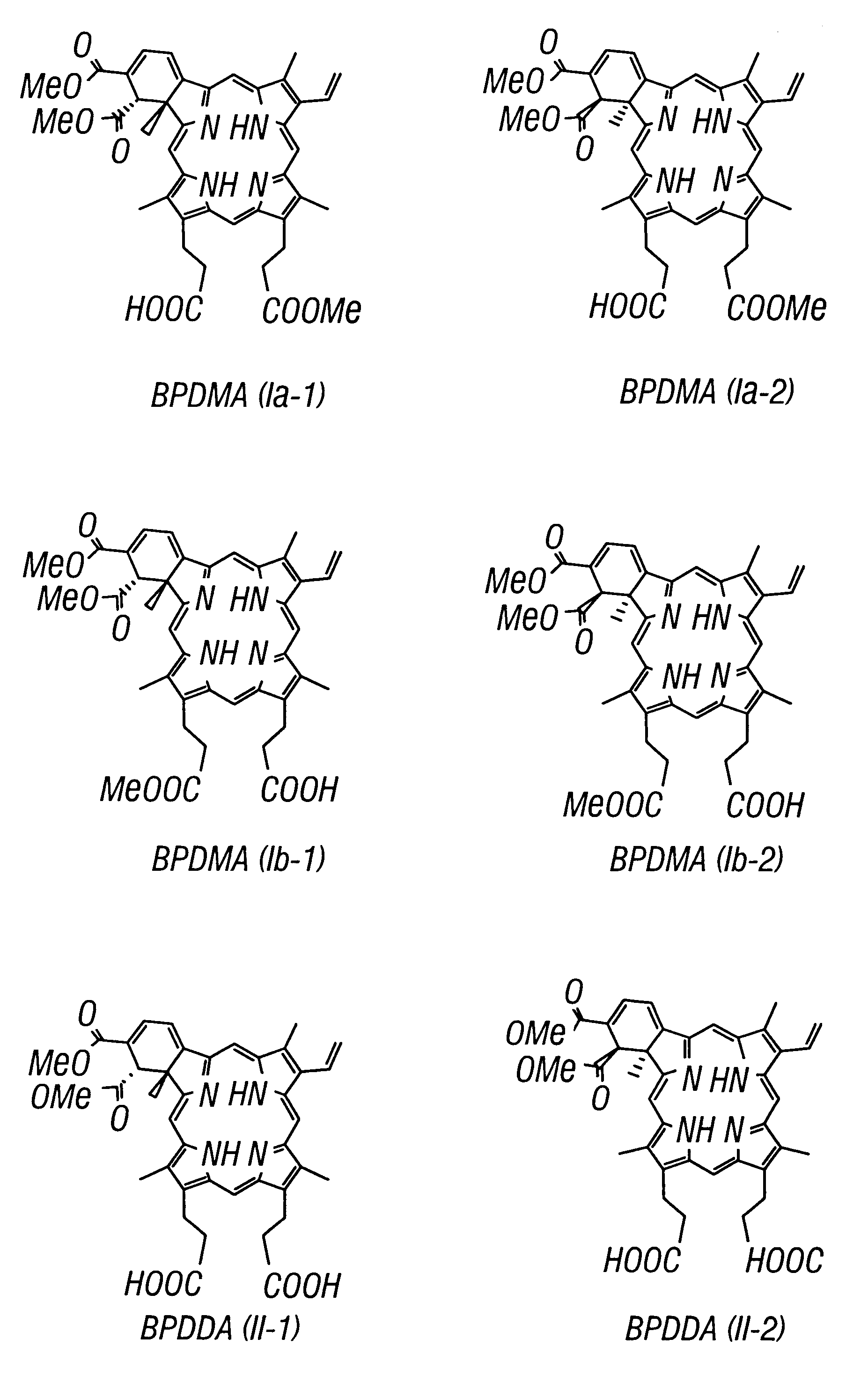
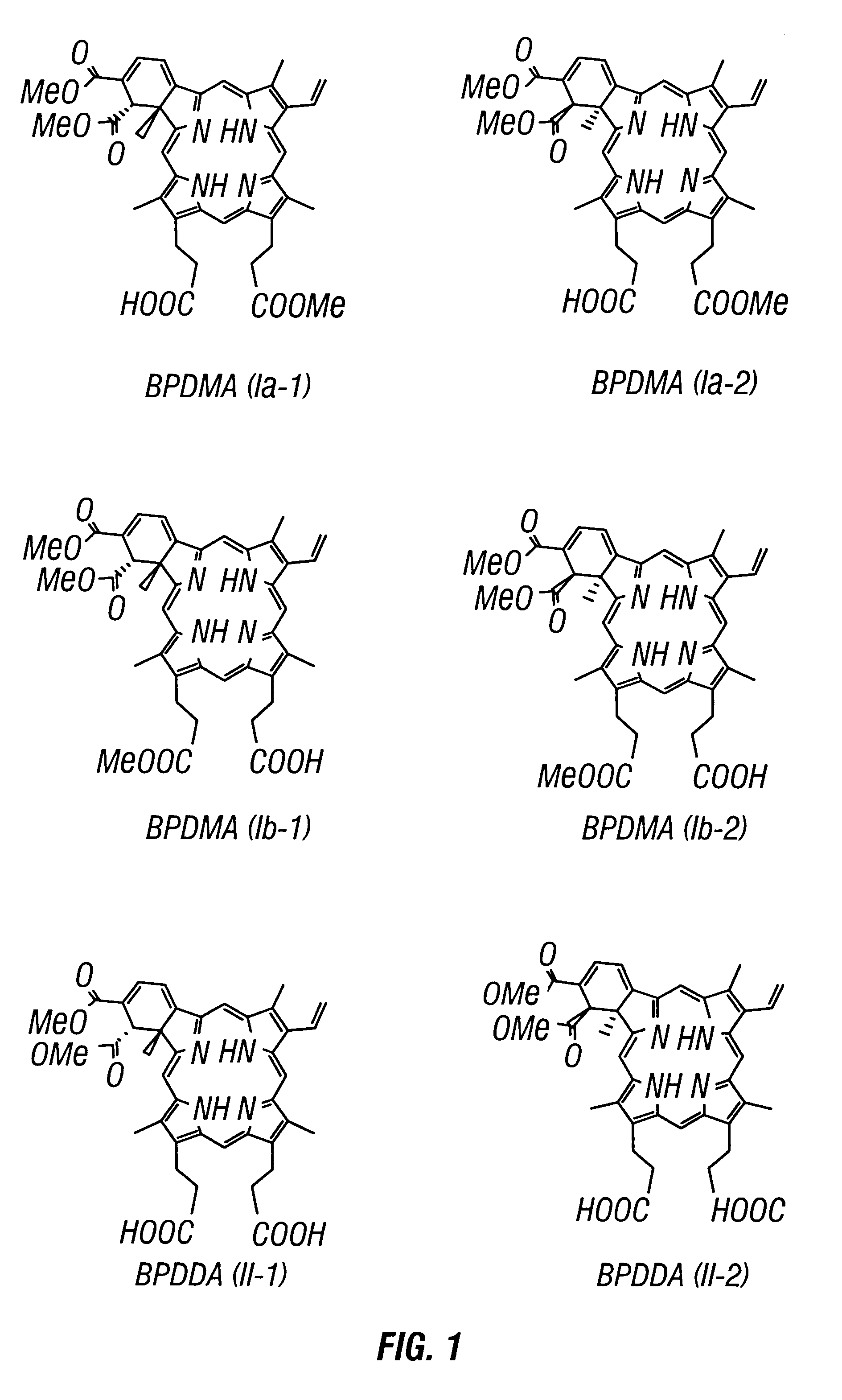
A method for the separation of benzoporphyrin derivative mono and diacid (BPD-MA, BPD-DA) enantiomers by Laser-Induced Fluorescence Capillary Electrophoresis has been developed. The limits of detection are 2.06x10-6 M, and the relative standard deviation for the separation was 2.90% to 4.64%. The BPD enantiomers can be quantitatively determined in the range of 10-2 to 10-5 mg mL-1. In comparison with HPLC, CE has better resolution and efficiency. This separation method was successfully applied to the BPD enantiomers obtained from a matrix of bovine serum and from liposomally formulated material as well as from studies with rat, dog and human microsomes.
Owner:THE UNIV OF BRITISH COLUMBIA
Antiviral compound preparation for fish
InactiveCN103372211AReduce dosageExpand insecticidal spectrumOrganic active ingredientsAntiparasitic agentsSulfur drugMicrosphere
The invention provides an antiviral compound preparation for fish. The compound preparation composition comprises microparticles containing macrolide anthelmintic, as well as two or more than two of bithionol, praziquantel, diflubenzuron, triflumuron, pyriproxyfen, sulfur powder, salicylanilide, organic phosphorus, benzimidazole, pyrethroid, triazine, tetramisole anthelmintic, sulfonamides, anticoccidial drugs and Chinese herbal medicine anthelmintic, wherein the microparticles containing macrolide anthelmintic are microspheres or microcapsules composed of biodegradable or non-biodegradable carrier materials and macrolide anthelmintic, and the content of the macrolide anthelmintic in the microparticles is 0.1-50%. The compound preparation provided by the invention can not only expand the insecticidal spectrum, but also effectively improve the anthelmintic activity and reduce toxicity so as to ensure safer use.
Owner:江苏海辰科技集团有限公司
Methods for treating patients with familial hypercholesterolemia
InactiveUS20170312359A1Reducing lipidExtended time intervalMetabolism disorderPharmaceutical delivery mechanismAntigenLipid lowering drug
The present invention provides methods for treating patients suffering from familial hypercholesterolemia, including both HeFH and HoFH. The methods of the invention provide for lowering at least one lipid parameter in the patient by administering a therapeutically effective amount of an antibody or antigen-binding fragment thereof that specifically binds to ANGPTL3 in combination with a therapeutically effective amount of a statin, a first lipid lowering agent other than a statin, and a second lipid lowering agent other than a statin. The first non-statin lipid lowering agent is an agent that inhibits cholesterol uptake (e.g. ezetimibe) and the second non-statin lipid-lowering agent is an inhibitor of microsomal triglyceride transfer protein (e.g. lomitapide). The combination therapy is useful in treating hypercholesterolemia, as well as hyperlipidemia, hyperlipoproteinemia and dyslipidemia, including hypertriglyceridemia, chylomicronemia, and to prevent or treat diseases or disorders, for which abnormal lipid metabolism is a risk factor, such as cardiovascular diseases.
Owner:REGENERON PHARM INC
Biological marker-earthworm internal organ p450 content measuring method
InactiveCN101210918AGuaranteed accurate quantificationAvoid Measuring InaccuraciesPreparing sample for investigationColor/spectral properties measurementsCytochrome P450Glycerol
The invention relates to cytochrome P450, specifically to a method for measuring the content of a biomarker-cytochrome P450 in earthworm viscera, which comprises the following steps of: (1) soaking a live earthworm in glycerol solution, and immediately dissecting and taking out the viscera thereof; (2) cleaning the viscera with cold saline water, transferring to a homogenization buffer, breaking cells with a glass tissue grinder, centrifuging with a ultra-speed centrifuge at low temperature (3DEG C) and 10,000g for 20min, centrifuging the supernatant at 150,000g for 90min, removing the supernatant, and collecting microsome sediment; and (3) dissolving the microsome sediment in a storage buffer, mixing, and measuring the total amount of cytochrome P450 and the protein content in the microsome suspension. The total amount of cytochrome P450 is quantified with sodium hydrosulfite-reduced CO difference spectra method. The invention has the advantages of simplicity, rapidness, low consumption, and accurate qualification and quantitation.
Owner:SHENYANG INST OF APPL ECOLOGY CHINESE ACAD OF SCI
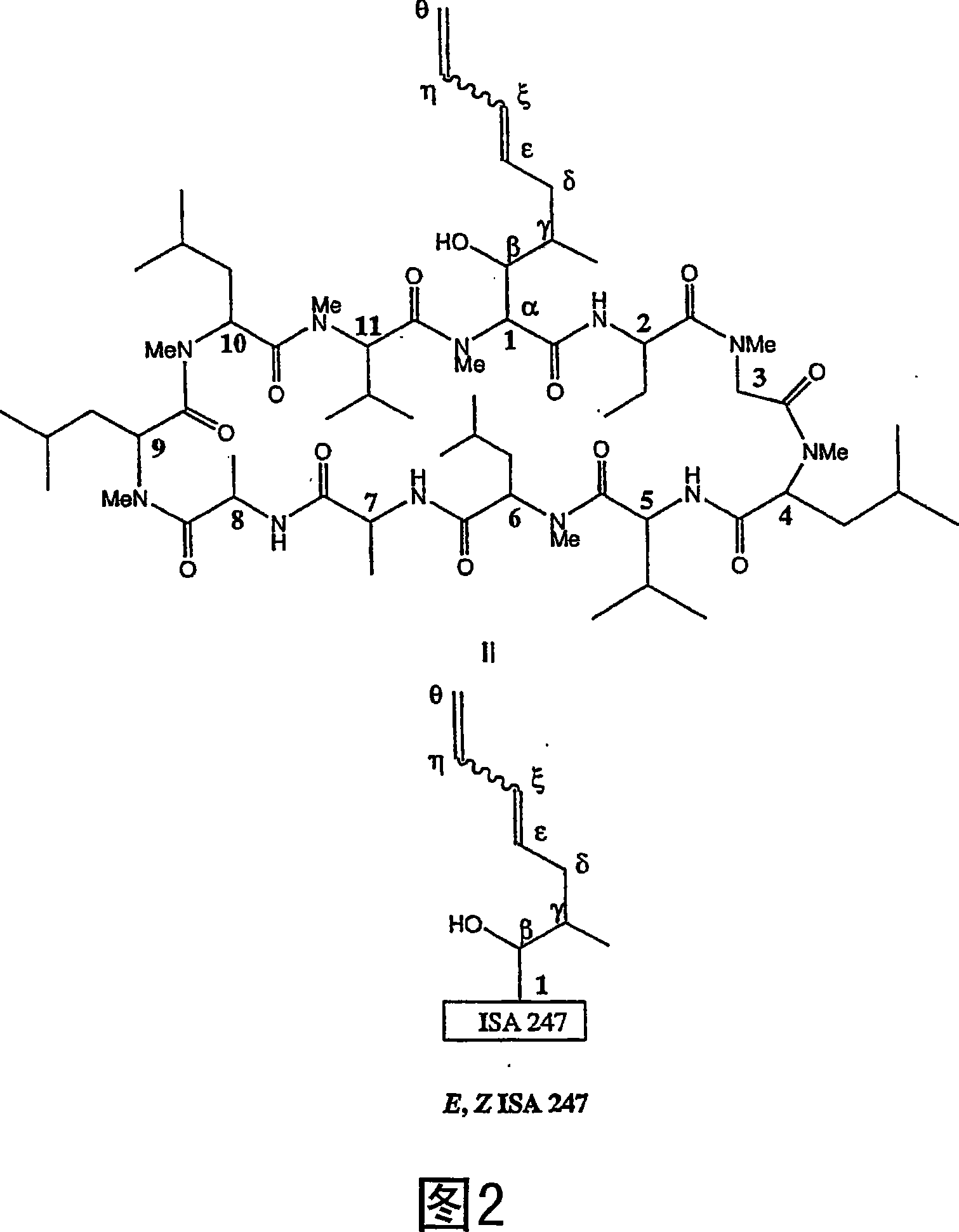
Metabolites of cyclosporin analogs
Isolated metabolites of the cyclosporine analog ISA247 are disclosed, including in vitro methods for their preparation. The metabolites comprise a chemical modification of ISA247, wherein the modification is at least one reaction selected from the group consisting of hydroxylation, N-demethylation, diol formation, epoxide formation, and intramolecular cyclization phosphorylation, sulfation, glucuronide formation and glycosylation. Methods of preparation include semi-synthetic methods, wherein metabolites of ISA247 are produced from the microsomal extracts of animal liver cells, or from cultures using microorganisms, and completely synthetic methods, such as chemically modifying the parent compound or isolated metabolites using organic synthetic methods.
Owner:伊素制药公司
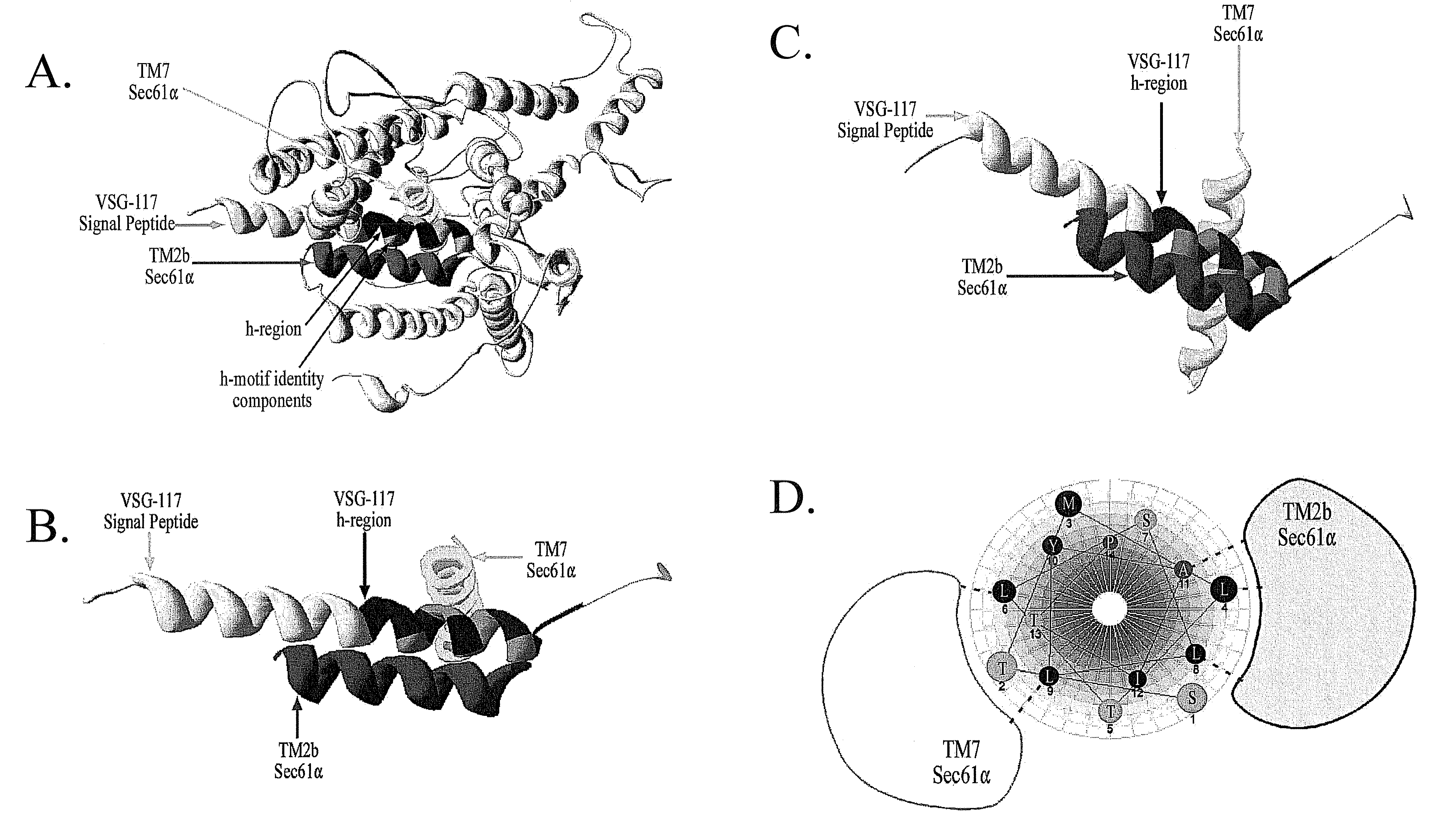
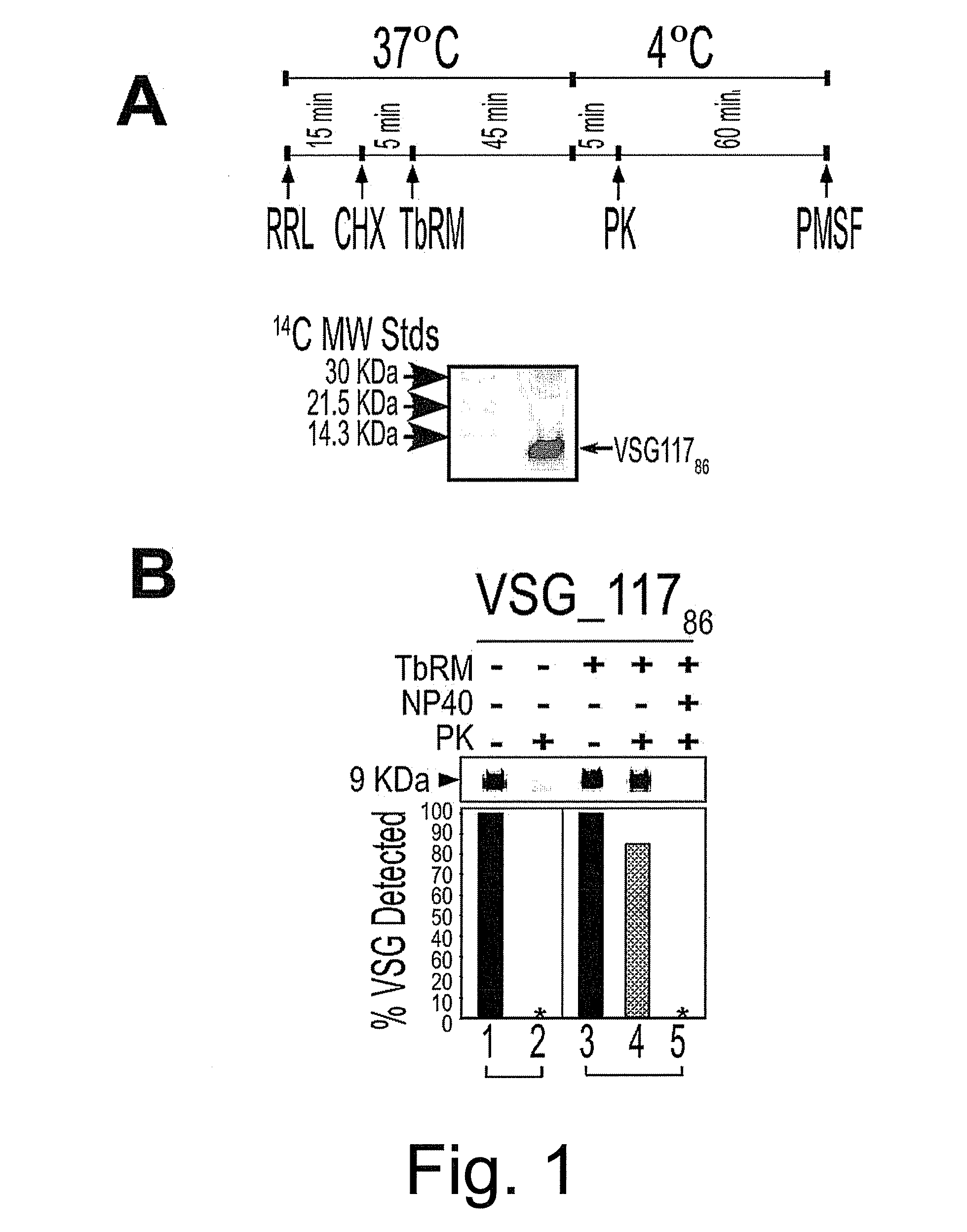
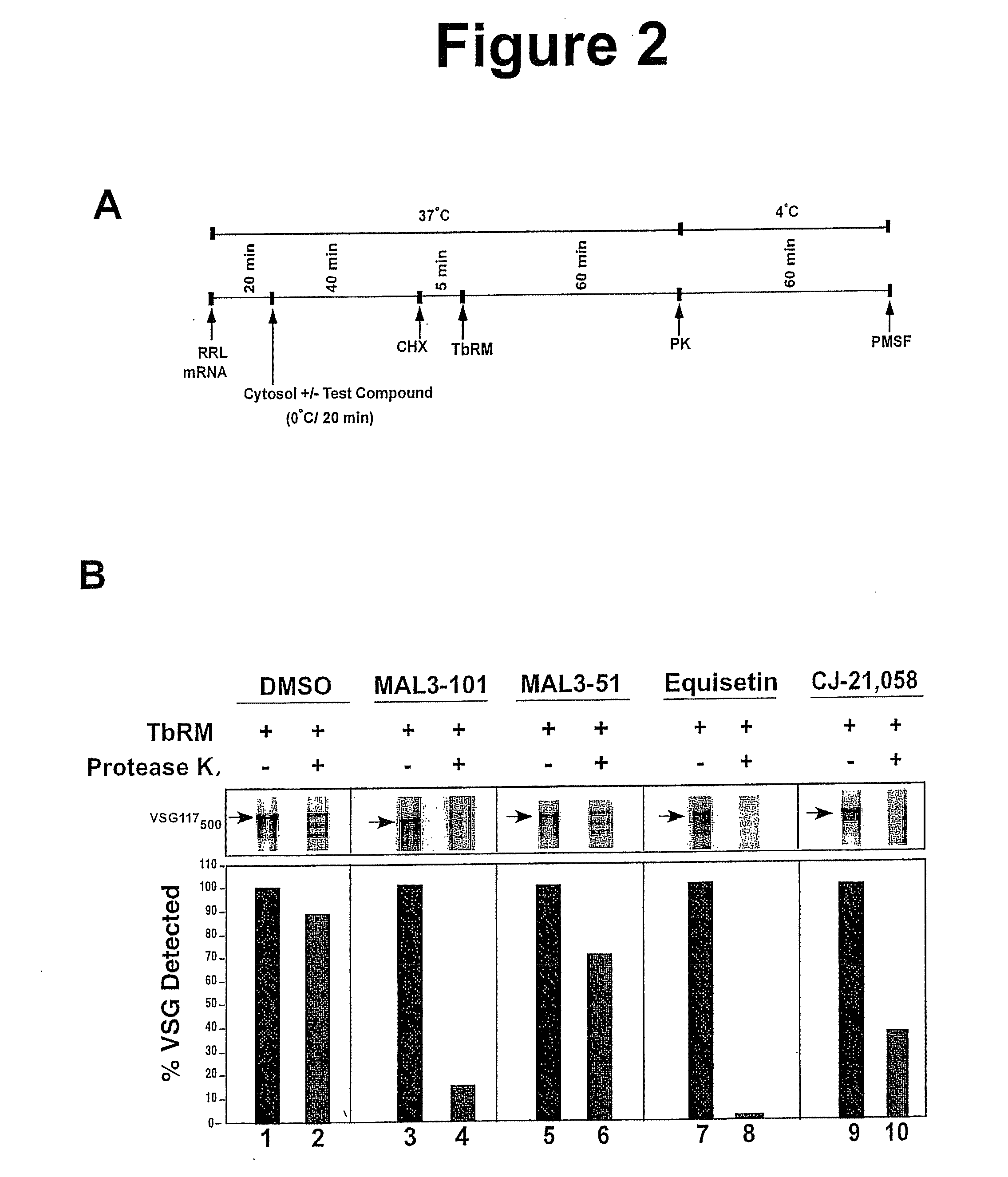
Trypanosome microsome system and uses thereof
The present invention provides cell free preparations of protozoan microsomes, wherein the cell free preparation of protozoan microsomes demonstrate the ability to translocate a protozoan polypeptide into the microsome, methods of preparing cell free preparations of protozoan microsomes, and methods of using cell free preparations of protozoan microsomes.
Owner:UNIV OF GEORGIA RES FOUND INC
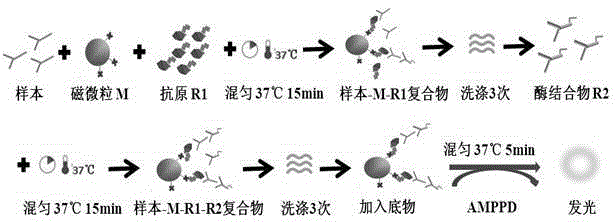
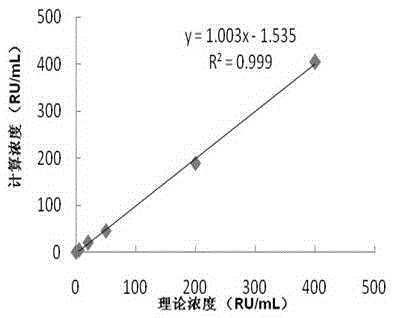
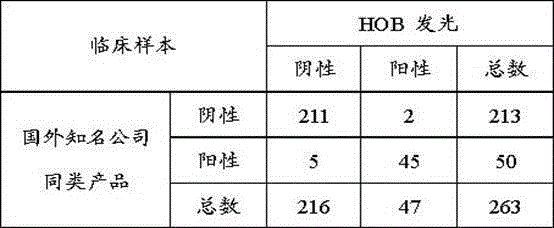
Anti-LKM (liver-kidney microsomal) 1 antibody detection kit and detection method
The invention discloses an anti-LKM (liver-kidney microsomal) 1 antibody detection kit and a detection method. The kit comprises biotinylation LKM-1 antigen, a calibrator, a quality control material, an AP (alkaline phosphatase)-anti-human IgG (intravenous gamma globulin) antibody, a magnetic particle reagent, a chemiluminescent substrate and a cleaning liquid. According to the detection method, the reagent in the kit is used for detection of the sample. With the adoption of the scheme, a full-automatic magnetism particulate chemiluminescent detection method is adopted for selection of an alkaline phosphatase AP-adamantane AMPPD system, and the kit has the advantages of good stability, high sensitivity, good repeatability and the like; meanwhile, the detection time is greatly shortened, the total time required for one test is about 50 minutes, the operation is simple and convenient, and full-automatic detection is really realized.
Owner:SUZHOU HAOOUBO BIOPHARML
Compositions from Garcinia as Aromatase Inhibitors for Breast Cancer Chemoprevention and Chemotherapy
InactiveUS20090181110A1Reduce frequencyShorten the durationBiocideMicrobiological testing/measurementDietary supplementDepressant
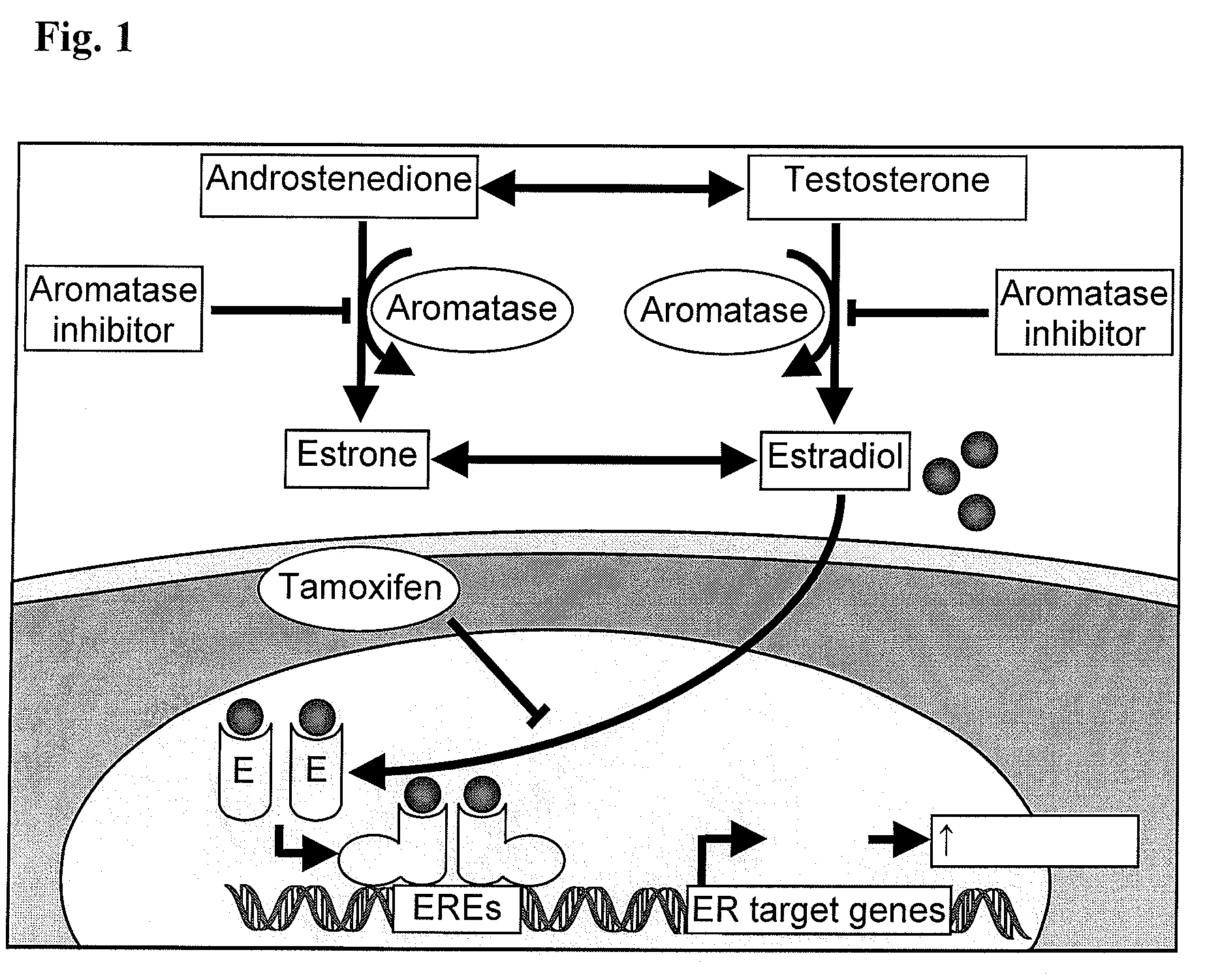
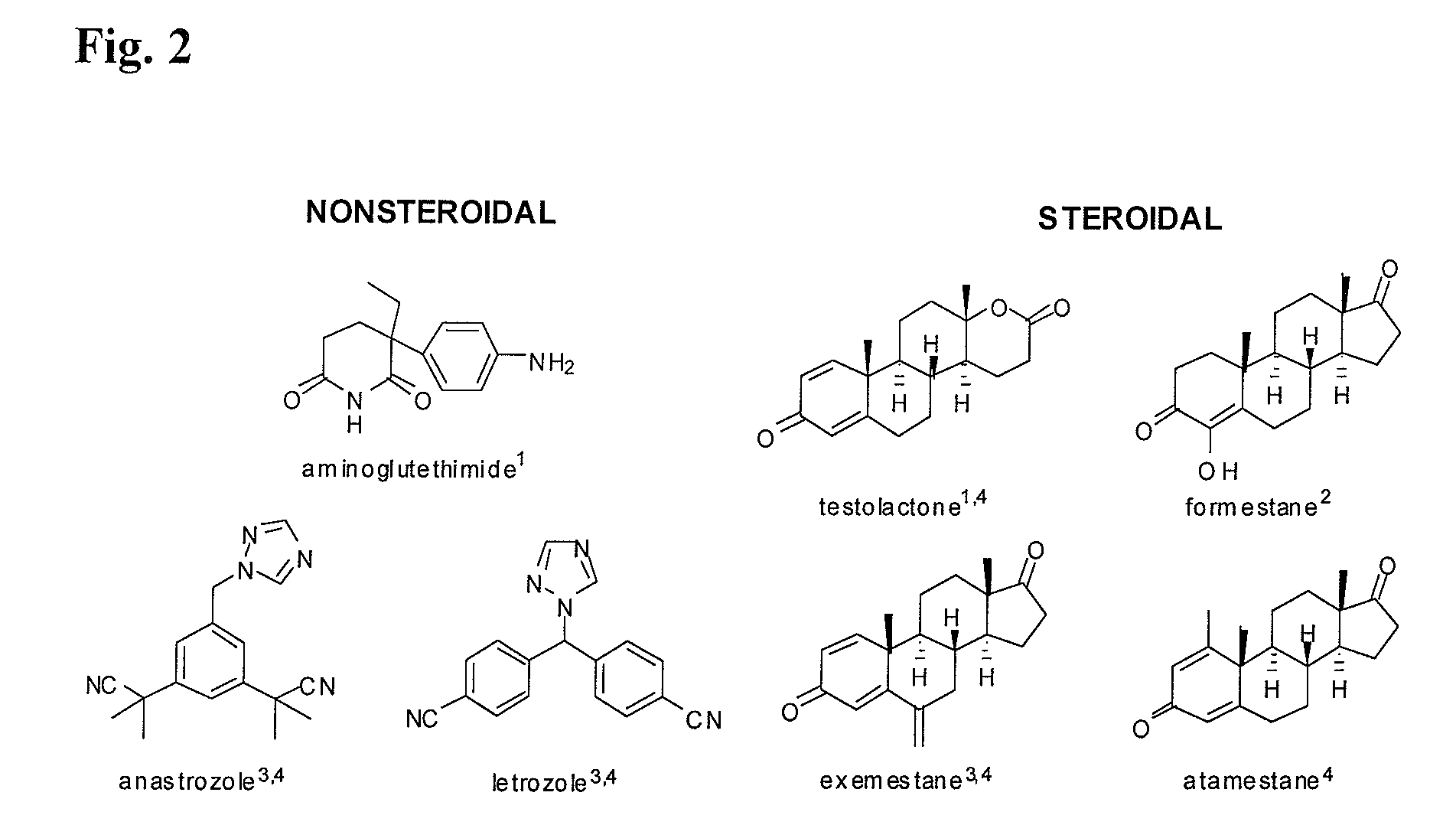
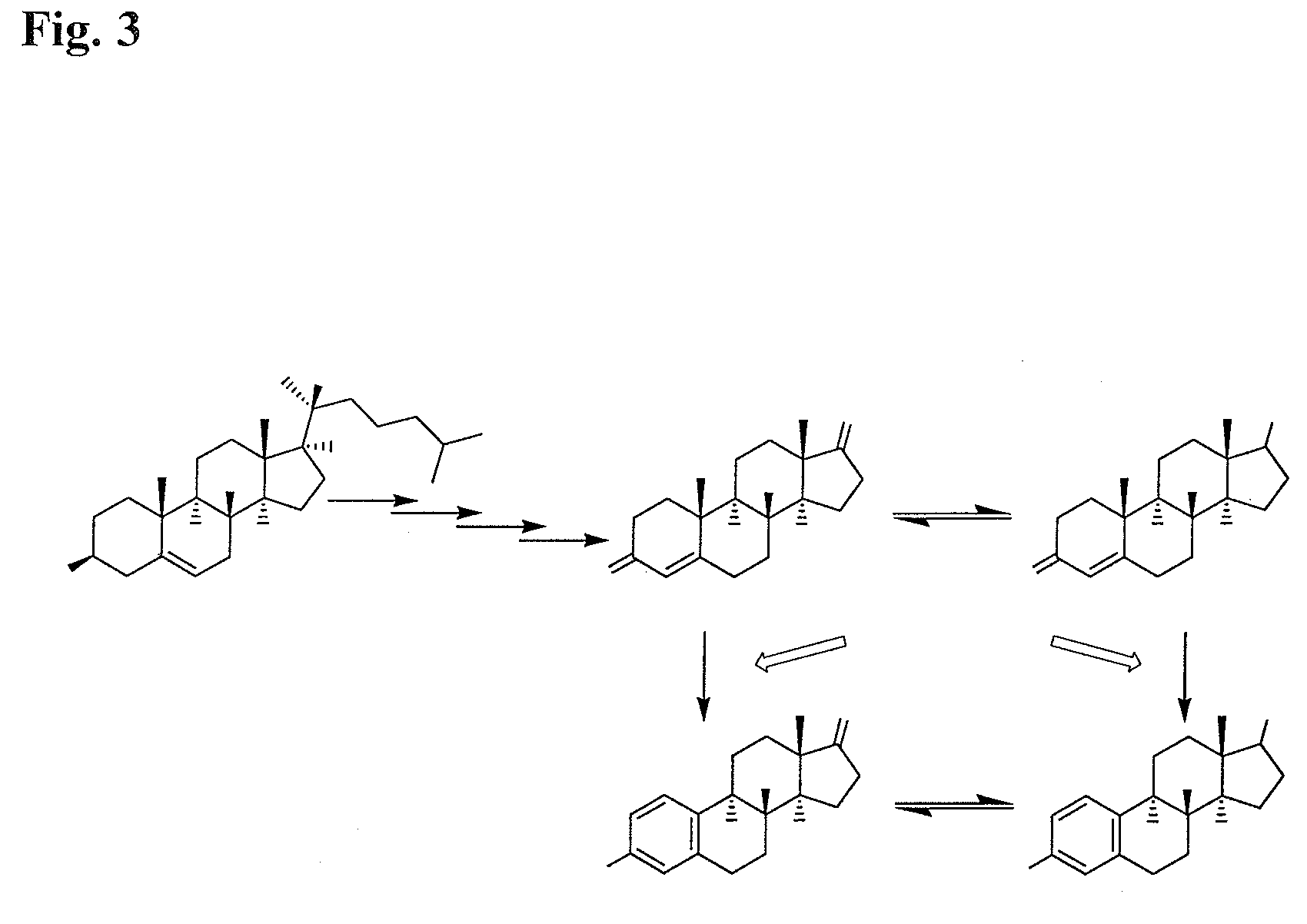
Aromatase inhibitors (AIs) are disclosed which are useful in the treatment and prevention of post-menopausal breast cancer. New AIs derived from natural products are disclosed that are evaluated for clinical utility for treating post-menopausal breast cancer and may also act as chemopreventive agents for preventing breast cancer. Several pure compounds demonstrated AI activity using a noncellular, enzyme-based microsomal and a cell-based aromatase assay. Correlations are made between structural classes with levels of aromatase inhibition. The disclosure may be utilized to direct synthetic modification of natural product scaffolds to enhance aromatase inhibition or to standardize botanical dietary supplements for increased aromatase inhibition activity.
Owner:THE OHIO STATE UNIV RES FOUND
Subcellular structure characteristic N-linked carbohydrate chain and application thereof
ActiveCN105651853AImplement indirect detectionEasy to detectMaterial analysis by electric/magnetic meansDetection of post translational modificationsBiologySubcellular structure
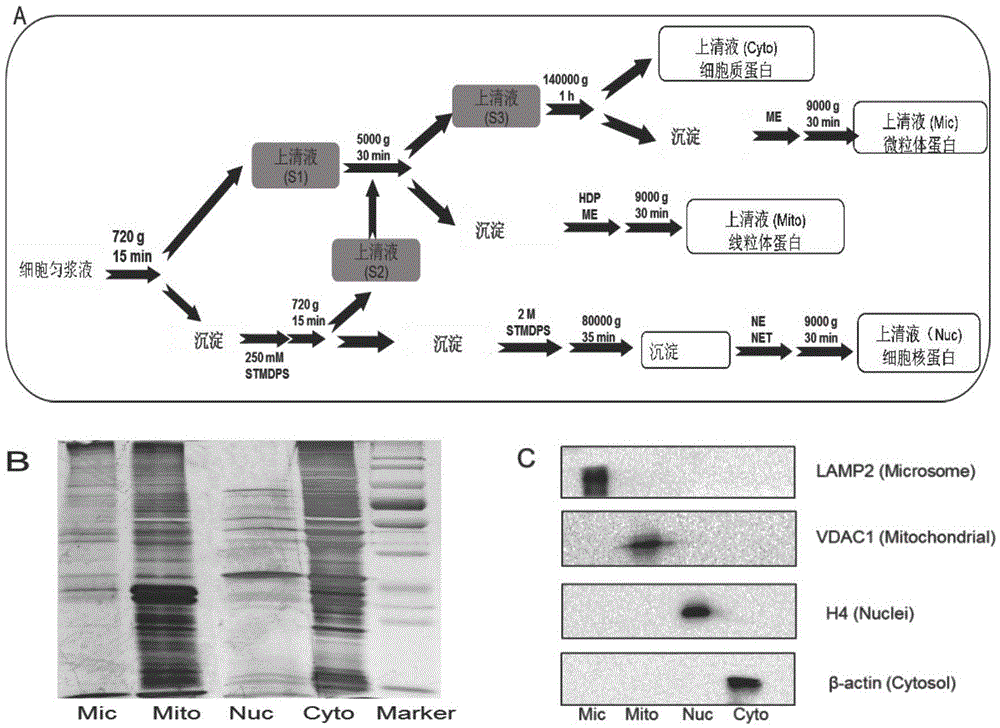
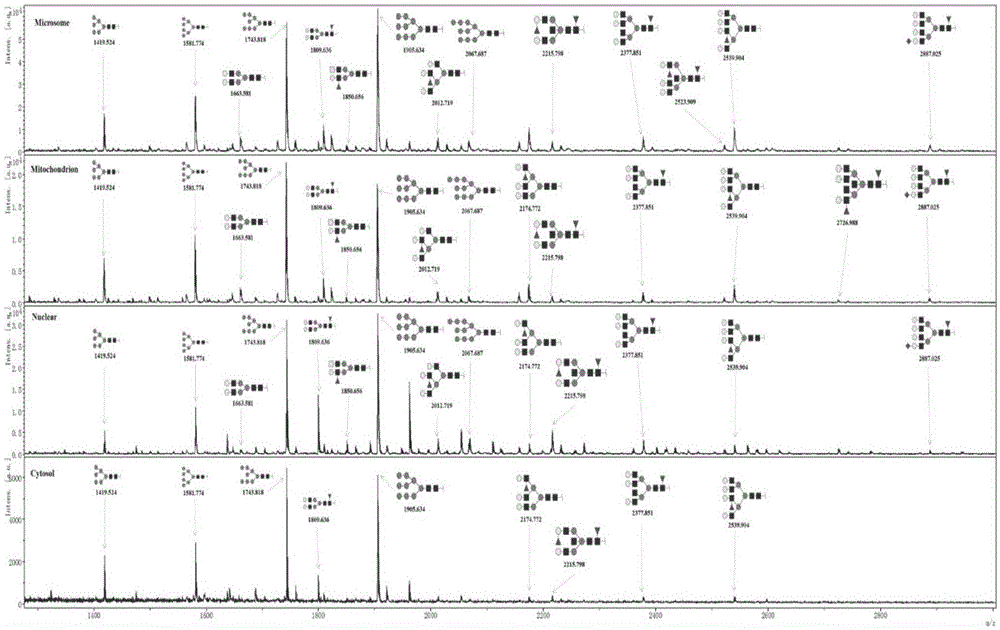
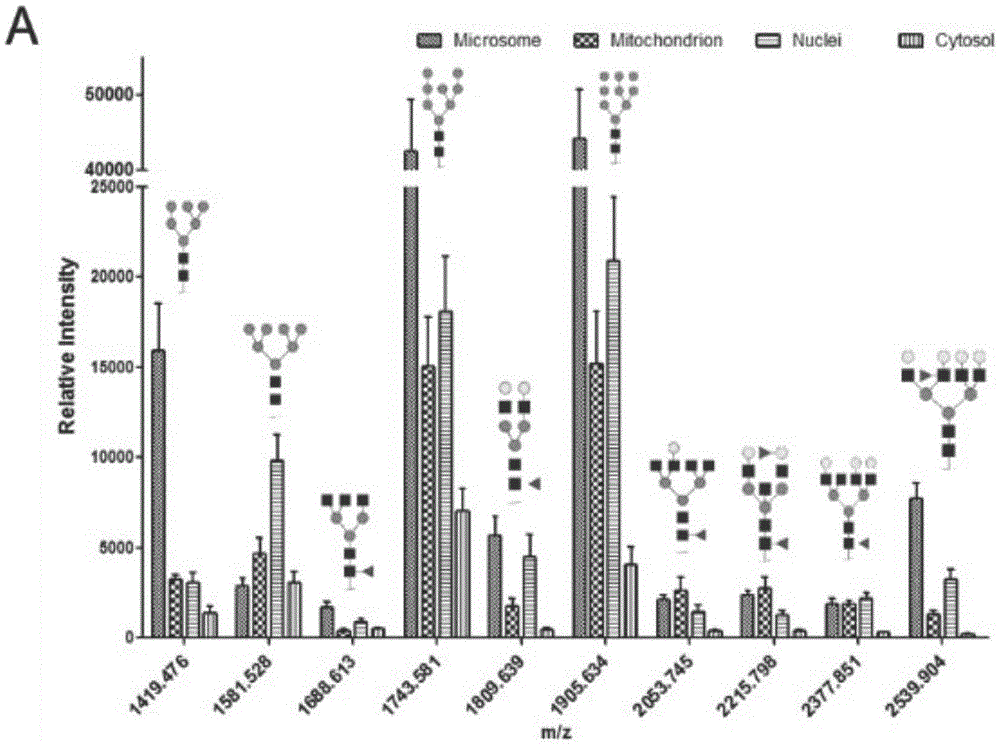
The invention discloses a subcellular structure characteristic N-linked carbohydrate chain and the application thereof and belongs to the technical field of glycobiology. Bladder YTS1 cells are used as the experiment material, the protein components of four subcellular structures (microsome, chondriosome, cell nucleus and cytoplasm) are obtained with the differential centrifugation method, and subcellular structure difference carbohydrate chain distribution is displayed through the change of the type and structure of subcellular structure protein galactosylated modification identified through a united lectin chip and a mass spectrum (PS) respectively. Subcellular structure characteristic N-linked glycan of ten kinds of human bladder YTS1 cells is obtained. By means of characteristic N-linked glycan, indirect detection and qualitative or quantitative detection of other target substances in corresponding subcellular structures can be achieved.
Owner:上海微江生物技术有限公司
Compounds
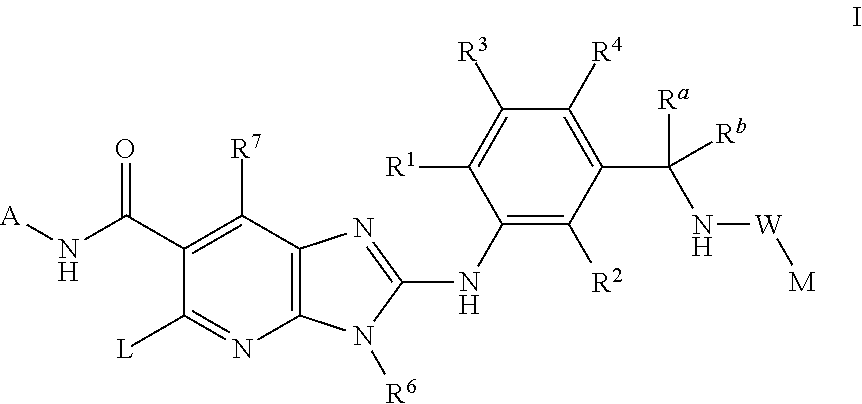
This invention relates to compounds of formula Itheir use as inhibitors of the microsomal prostaglandin E2 synthase-1 (mPGES-1), pharmaceutical compositions containing them, and their use as medicaments for the treatment and / or prevention of inflammatory diseases and associated conditions. A, L, M, W, R1, R2, R3, R4, R6, R7, Ra, Rb have meanings given in the description.
Owner:BOEHRINGER INGELHEIM INT GMBH
Determination of biological tag-earthworm in-vivo P450 content
InactiveCN1746661ALow costEfficient removalPreparing sample for investigationColor/spectral properties measurementsIce waterMicroparticle
A method for determining P450 content in biological tag ¿C earthworm body includes soaking earthworm living body in glycerin then moving it into salt solution for breaking it to be pieces, moving broken matter into homogenizing solution for homogenizing of 30 ¿C 50 second at 6000 ¿C 8000 rpm, then centrifugalizing homogenized matter for 20 ¿C 30 min at 12000 ¿C 15000 g under 3 ¿C 5 Deg C.; removing off supernatant then centrifugalizing for 90 ¿C 120 min at 120000 ¿C 150000g, discarding supernatant and putting particles in ice water, removing off red matter; dissolving particle in buffer solution and taking out 1 ¿C 2 mI for detecting protein content as the rest for being used to determine P450 content .
Owner:SHENYANG INST OF APPL ECOLOGY CHINESE ACAD OF SCI
Method for assaying compounds or agents for ability to decrease the activity of microsomal prostaglandin E synthase or hematopoietic prostaglandin D synthase
InactiveCN1675543ACompound screeningApoptosis detectionProstaglandin-E SynthasesProstaglandin product
Provided herein is a novel and useful method for evaluating the ability of compounds or agents to decrease the activity of microsomal prostaglandin E synthase or hematopoietic prostaglandin D synthase to produce their respective prostaglandin products.
Owner:AVENTIS PHARMA INC
Combinations of MTP Inhibitors with Cholesterol Absorption Inhibitors or Interferon for Treating Hepatitis C
InactiveUS20100226886A1Reduction in hepatitis C viral loadShort treatment timeBiocidePeptide/protein ingredientsActive agentMicrosomal triglyceride transfer protein
The invention is directed in part to methods for treating and / or controlling hepatitis C in a patient. The methods are directed in part to combination therapies using a microsomal triglyceride transfer protein (MTP) inhibitor (for example, AEGR-733 and implitapide) and at least one other active agent.
Owner:AEGERION PHARM INC
Method for obtaining wire-worm microsome amino peptidase gene and use thereof
InactiveCN101434951AImprove immune activityEasy accessHydrolasesAntiparasitic agentsTotal rnaStructural analysis
The invention provides a method for obtaining a haemonchus contortus (H.contortus) microsomal aminopeptidase gene which comprises the steps of total RNA extraction, H11 cDNA sequence clone, homology comparison and amino acid sequence analysis of H11 cDNA sequence as well as H11 genome sequence clone and structural analysis. By referring to the H11 gene cDNA sequence, the method can conveniently, completely and accurately obtain H11 genome sequence, can analyze the H11 mRNA transcription characteristic by analyzing the H11 genome sequence, and provides reliable background material and data for developing H.contortus vaccines. The haemonchus contortus (H.contortus) microsomal aminopeptidase gene that is provided by the invention can be applied to the subunit vaccine preparation that uses recombinant proteins.
Owner:ZHEJIANG UNIV
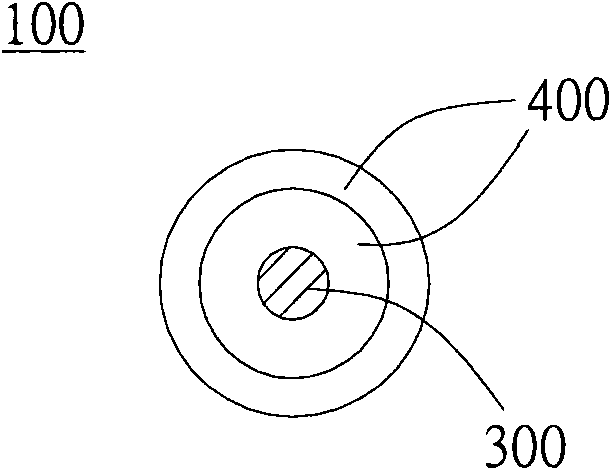
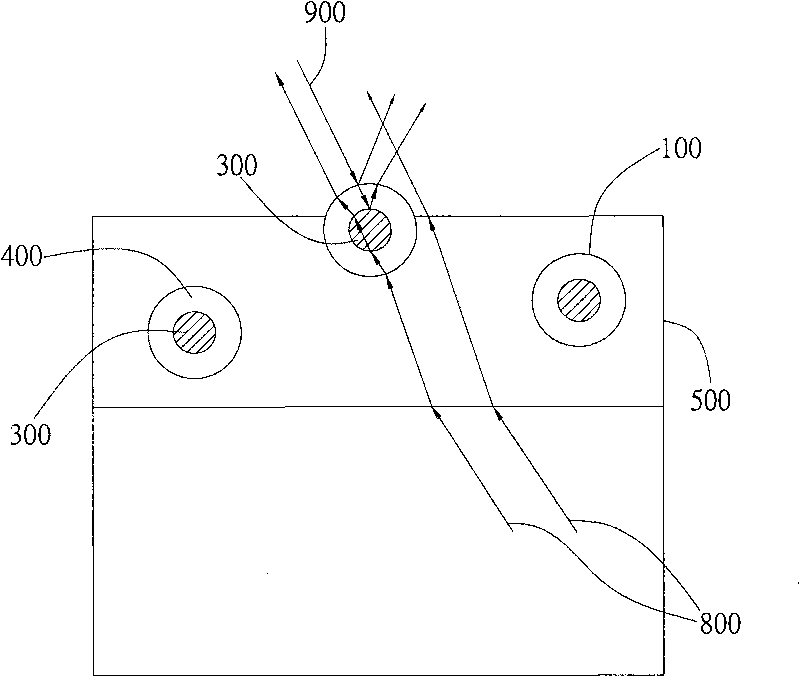
Multilayer microsome and anti-glare film thereof
InactiveCN101713837AReduce manufacturing costWith anti-glare functionDiffusing elementsSynthetic resin layered productsRefractive indexMicrosome
The invention relates to a multilayer microsome and an anti-glare film formed by applying the multilayer microsome. The anti-glare film contains transparent resin and a plurality of multilayer microsomes, wherein the multilayer microsomes are evenly distributed in the transparent resin. Since the multilayer microsomes consist of at least two layers of different transparent substances, the phenomenon of different light refractive indexes can be generated, and light scattering and refraction are caused further.
Owner:DAXON TECH INC
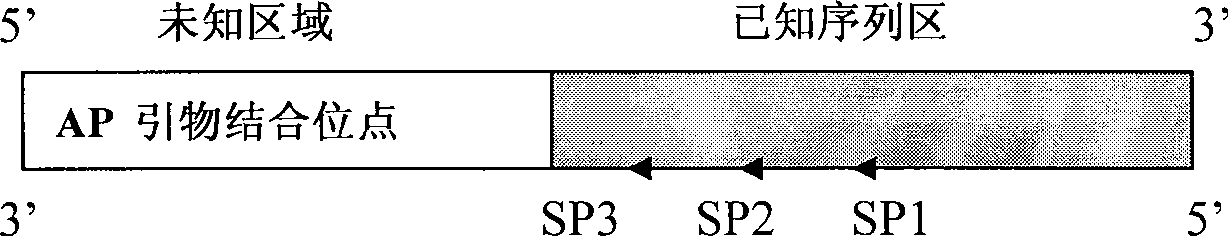
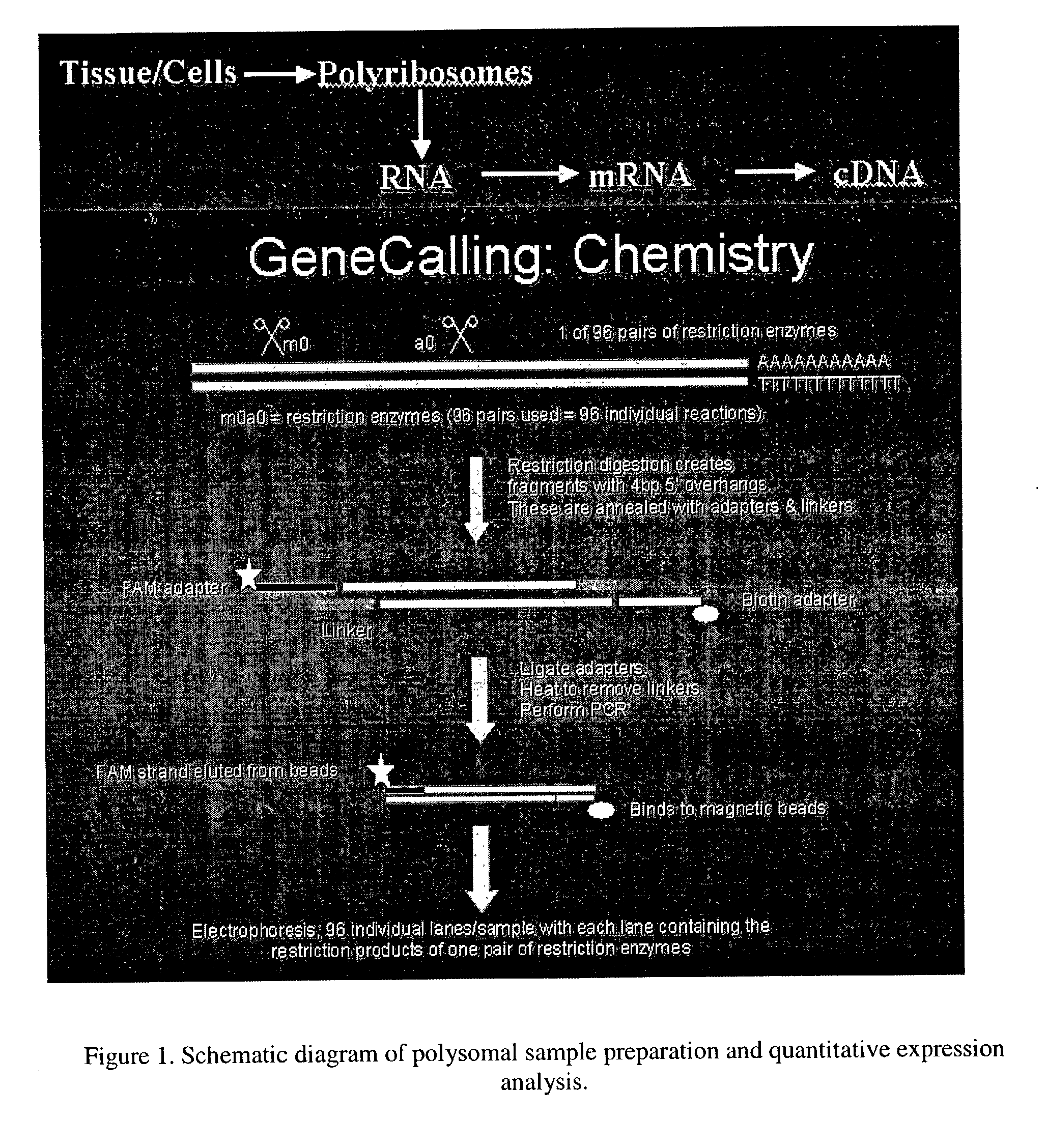
Method for analyzing a nucleic acid
InactiveUS20070178452A1Microbiological testing/measurementBiological testingNucleic acid detectionSample sequence
Disclosed is a method in which DNA sequences derived from microsome-associated mRNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information
Owner:BOUFFARD PASCAL +8
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com