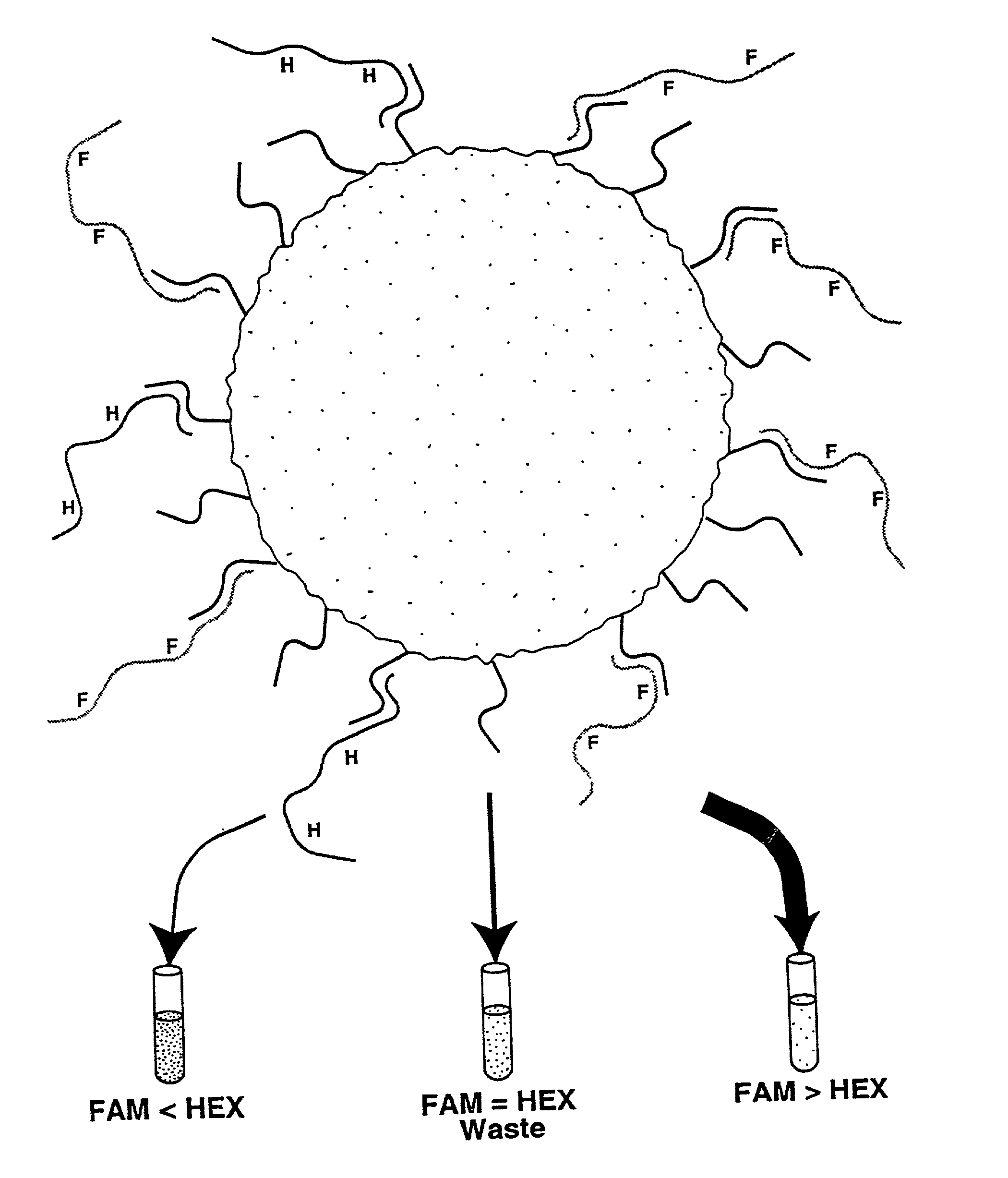
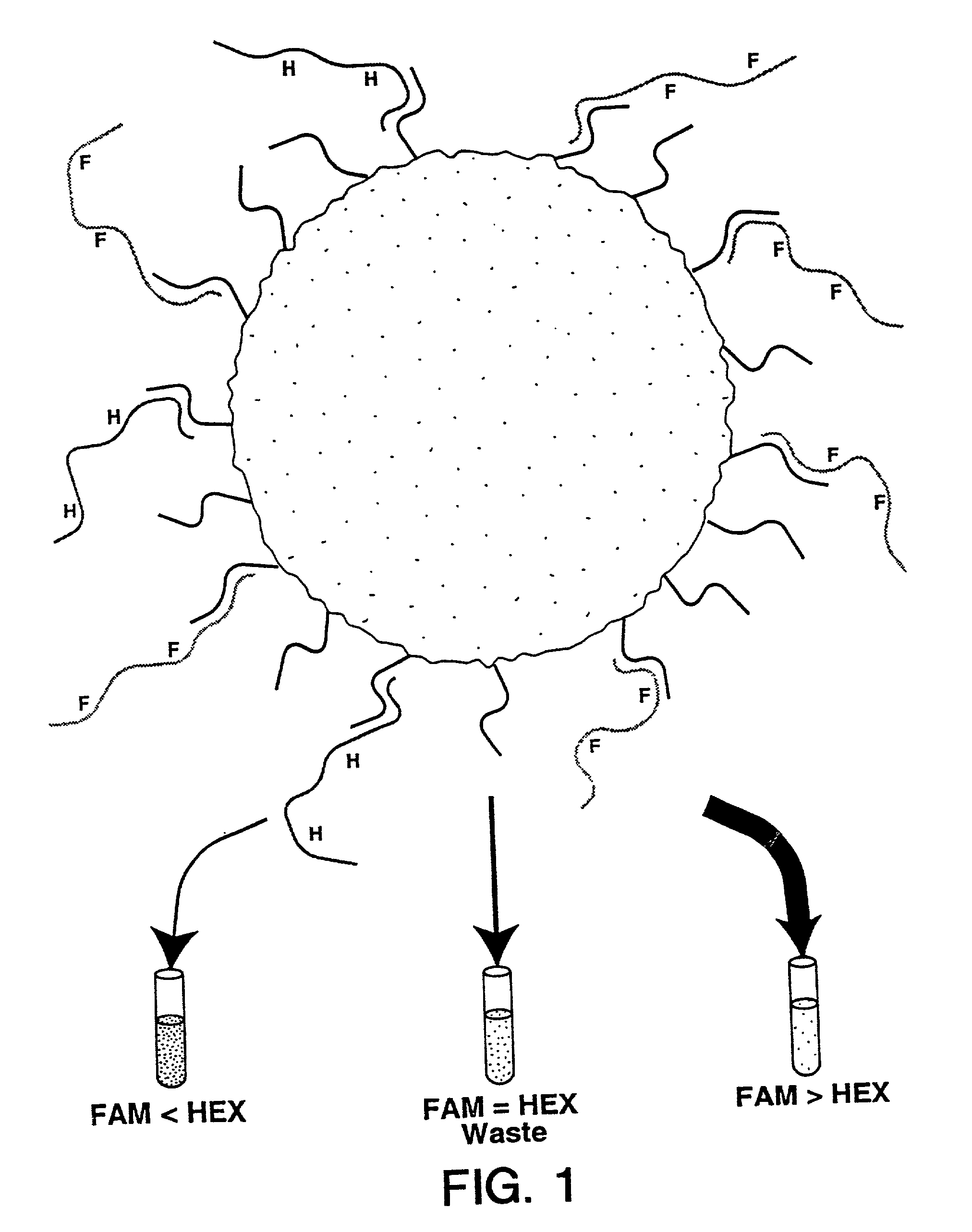
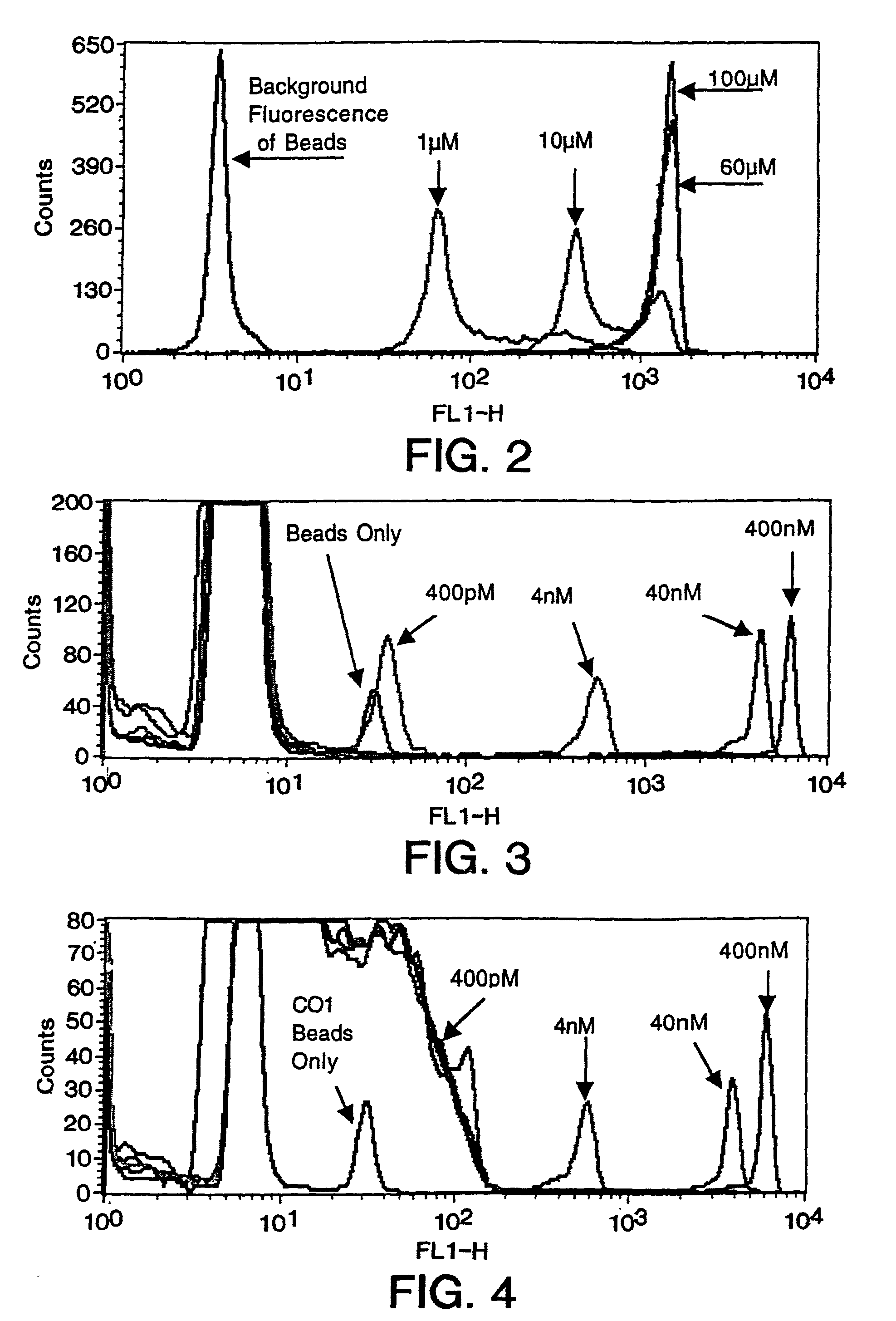
Methods for negative selections under solid supports
a technology of solid supports and negative selections, applied in the field of quantitative and isolation, can solve the problems of slow and expensive sequencing, limited genomic sequencing, and laborious process of positional gene cloning,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
first embodiment
[0091] In a first embodiment, glass beads are treated with 3-glycidoxypropyltrimethoxysilane to generate a terminal epoxide conjugated via a linker to Si atoms on the glass. In a second step, the epoxide is opened with either water or a diol to generate alcohols. Maskos and Southern, 1992, Nucleic Acids Research 20:1679-1684. The resulting siloxane linkage is relatively stable to base hydrolysis. Glass beads are a necessary starting material to produce hydroxyl groups suitable to begin cycles of phosphoramidite chemistry in a conventional automated DNA synthesizer. In some preferred applications, commercially available controlled-pore glass (CPG) or polystyrene supports are employed as beads. Such supports are available with base labile linkers and initial nucleosides attached, by, e.g., Applied Biosystems (Foster City, Calif.). Alternatively, non-porous glass beads, e.g., Ballotini spheres are employed (Maskos and Southern, 1992, Nucleic Acids Research 20:1679-1684).
second embodiment
[0092] In a second embodiment, the linkage is created by the reaction of primary amines with phosphoramidite nucleotides to produce a base-stable linkage. Pon et al., 1988, Biotechniques 6:768-775. In the first step of the reaction an N-P linkage is formed due to nucleophilic attack by nitrogen on phosphorus. This linkage is oxidized in a subsequent step to the phosphoramidate, a stable chemical linkage. Beads that are functionalized with surface primary amines can be obtained from commercial sources.
third embodiment
[0093] In a third embodiment, the capture oligonucleotides are attached to the bead via a phosphodiester bond generated by standard phosphoramidite synthesis utilizing the attack of bead-linked hydroxyl oxygens on the nucleotide phosphorus to produce a phosphodiester bond, following oxidation with molecular iodine. Others have utilized this reaction to generate stable linkages (e.g., Needels et al, 1993, Proc. Natl. Acad. Sci. U.S.A. 90:10700-10704). The key step is the derivatization of appropriate beads such that they contain significant numbers of hydroxyl functional groups on their surface. It is possible to purchase such functionalized beads from a variety of commercial sources; the capture oligonucleotides may be synthesized chemically on the surface of these functionalized beads.
[0094] Generally, standard synthesis chemistries are used, such as phosphoramidite chemistry, as disclosed in Beaucage and Iyer, 1992, Tetrahedron 48:2223-2311, Molko et al., U.S. Pat. No. 4,980,460; ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Gene expression profile | aaaaa | aaaaa |
| Fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com