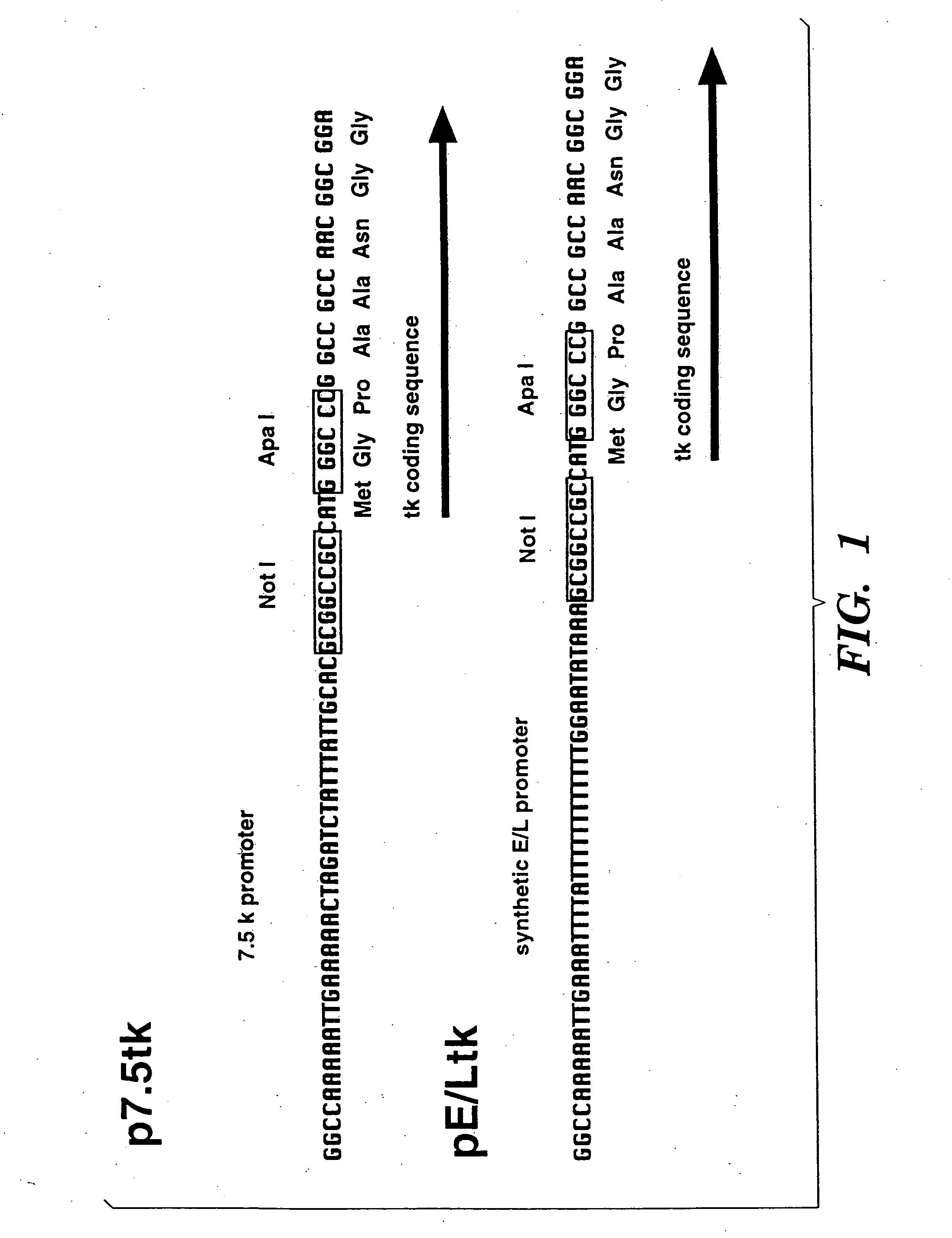
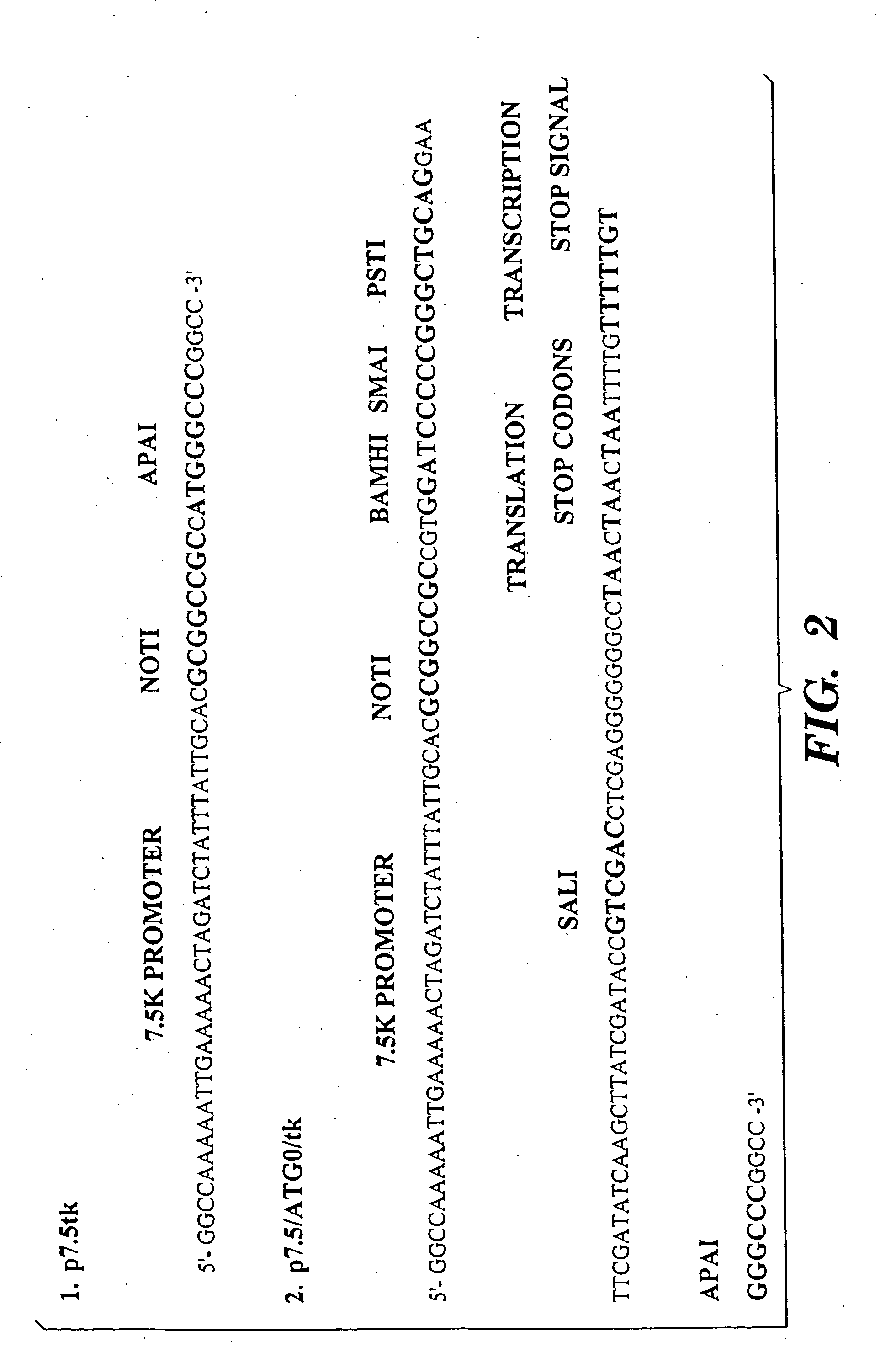
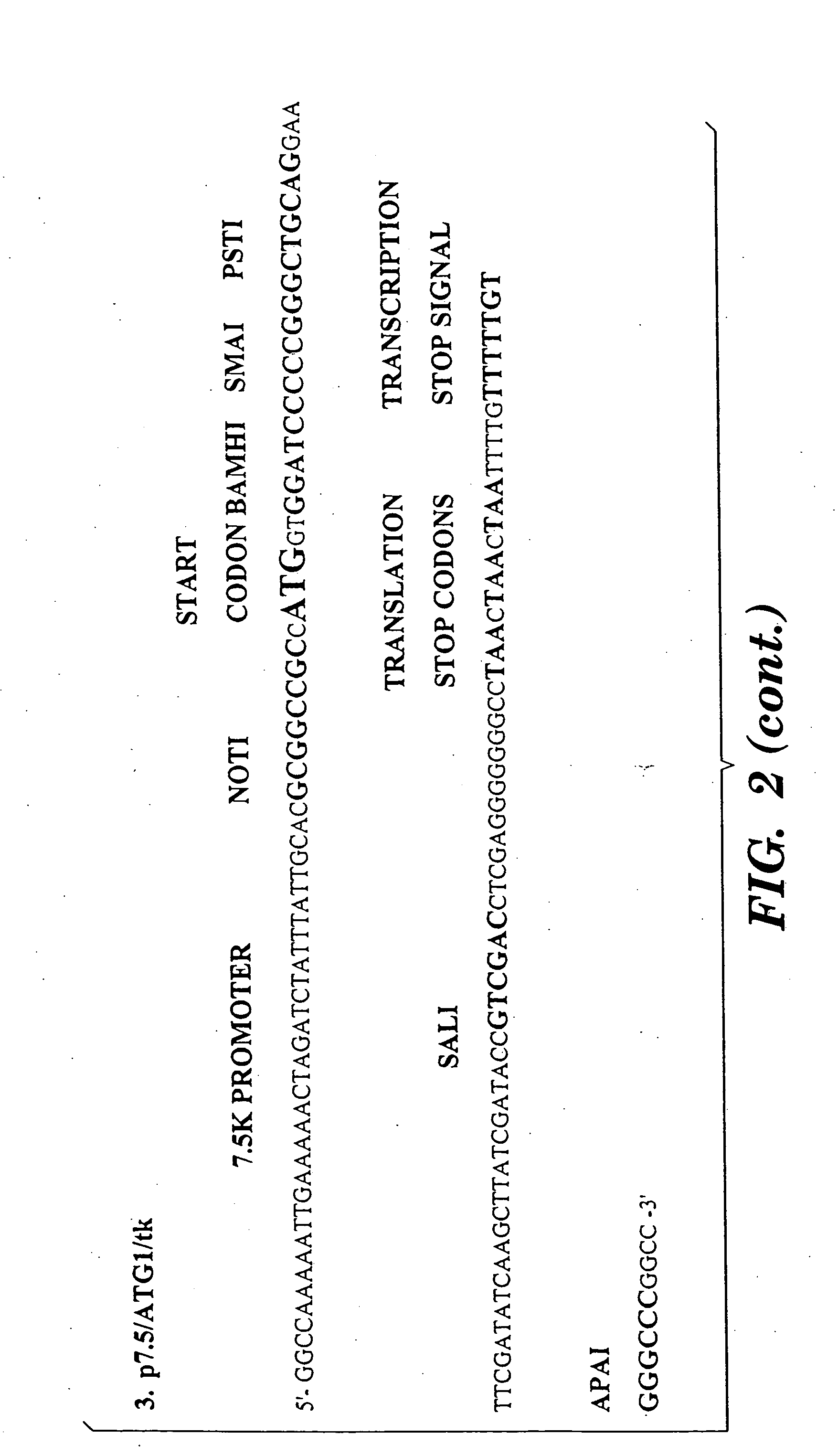
T cells specific for target antigens and methods and vaccines based thereon
a technology of t cells and target antigens, applied in the field of t cells specific for target antigens and methods and vaccines based thereon, can solve the problems of reducing false positives, reducing the resolution of false positives, and achieving limited success
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
Induction of Cytotoxic T Cells Specific for Human Tumors in HLA and Human CD8 Transgenic Mice
[0150] In this example, HLA and human CD8 transgenic mice were tolerized with a non-tumorigenic, immortalized normal human cell line that does not express costimulator activity for murine T cells and were subsequently immunized with B7 (costimulator) transfected tumor cells derived by in vitro mutagenesis or oncogene transformation from that same normal cell line. The HLA transgene permits selection of a high affinity, HLA-restricted T cell repertoire in the mouse thymus. In addition, a human CD8 transgene is required because murine CD8 does not interact efficiently with human class I MHC. Subsequent to immunization with B7 transfected tumor cells, splenic CD8+T cells are isolated and stimulated again in vitro in the absence of costimulation with non-tumorigenic, immortalized human cells. Two pathways of tolerance induction for antigens shared by the tumorigenic and non-tumorigenic cell line...
example 3
High-Throughput Strategy For Selection of DNA Recombinants From a Library That Encodes The Target Epitopes of Specific Cytotoxic T Cells
[0154] In this example, a model system was assayed to determine the level of enrichment that can be obtained through a procedure that selects for DNA recombinants that encode the target epitopes of tumor specific cytotoxic T cells.
[0155] Methods and Results
[0156] A specific vaccinia recombinant that encodes a well characterized ovalbumin peptide (SIINFEKL) (SEQ ID NO:10) was diluted with non-recombinant virus so that it constituted either 0.2%, 0.01%, or 0.001% of viral pfu. This ovalbumin peptide is known to be processed and presented to specific CTL in association with the murine class I MHC molecule H-2 Kb. An adherent monolayer of MC57G cells that express H-2 Kb were infected with this viral mix at m.o.i.=1 (approximately 5.times.10.sup.5 cell / well). MC57G cells do not themselves express ovalbumin peptide, but do express H-2 Kb, which allows the...
example 4
[0162] Identification of Potential Tumor-specific Antigens That are Differentially Expressed in Tumors
[0163] Identification of genes that are differentially expressed in human tumors, cancers, or infected cells could facilitate development of broadly effective human vaccines. Most methods for identification of differential gene expression are variations of either subtractive hybridization or the more recently described differential display technique.
[0164] Representational difference analysis (RDA) is a subtractive hybridization based method applied to "representations" of total cellular DNA (Lisitsyn, N. and N., M. Wigler, "Cloning the differences between two complex genomes," Science 259:946-951 (1993)). The differential display methods of Liang and Pardee (Science 257:967-971 (1992)) employ an arbitrary 10 nucleotide primer and anchored oligo-dT to PCR amplify an arbitrary subset of fragments from a more complex set of DNA molecules. As described below (Example 4), we have modifi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com