Flavivirus expression and delivery system
a technology of flavivirus and expression system, applied in the field of flavivirus expression and delivery system, can solve the problems of inability to glycosylate proteins, inefficient post-translational modification, and inability to cleave “prepro” or “prepro” sequences from proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cells.
[0099] BHK2 incubator.
Construction of the Replicons and Vectors.
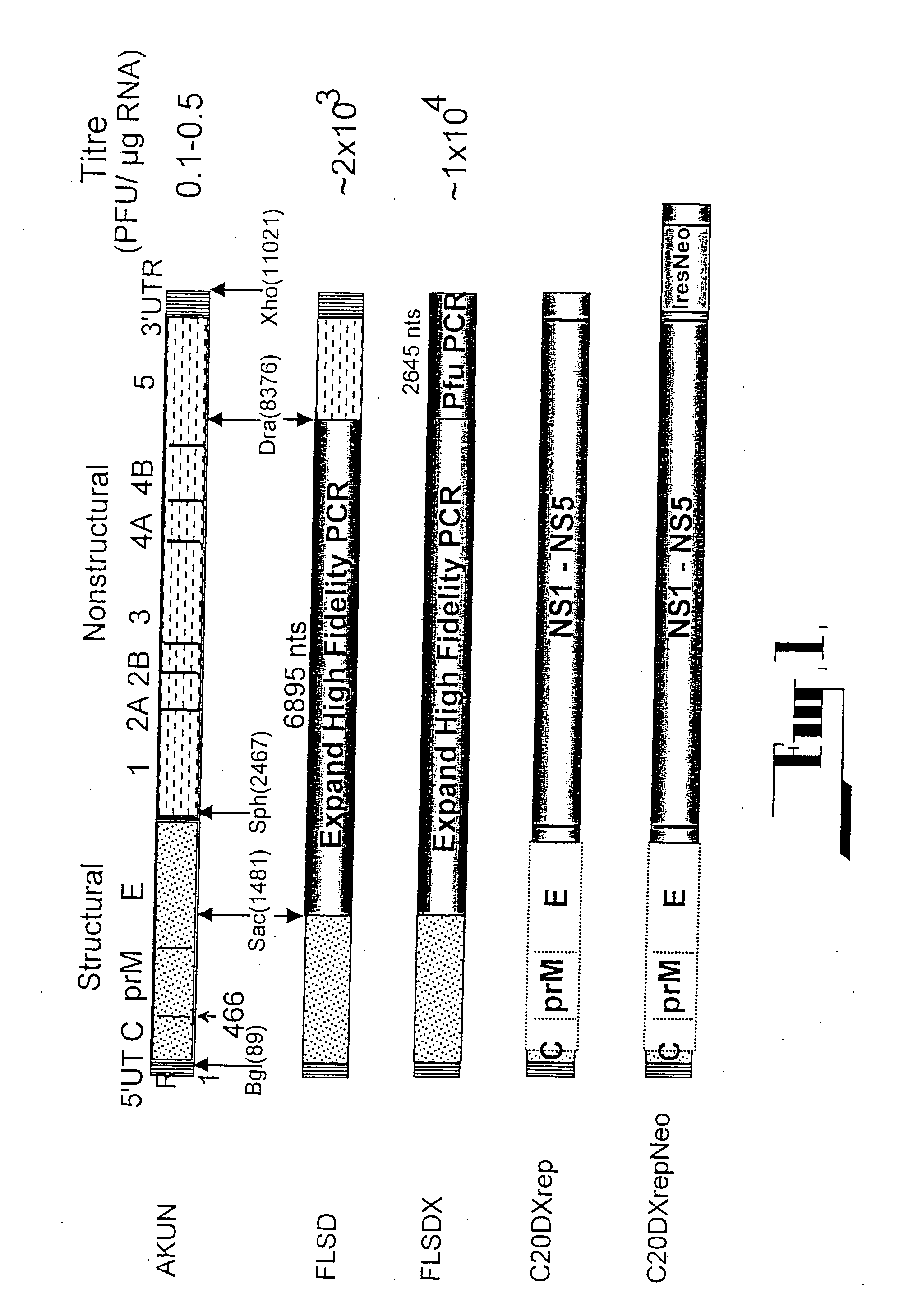
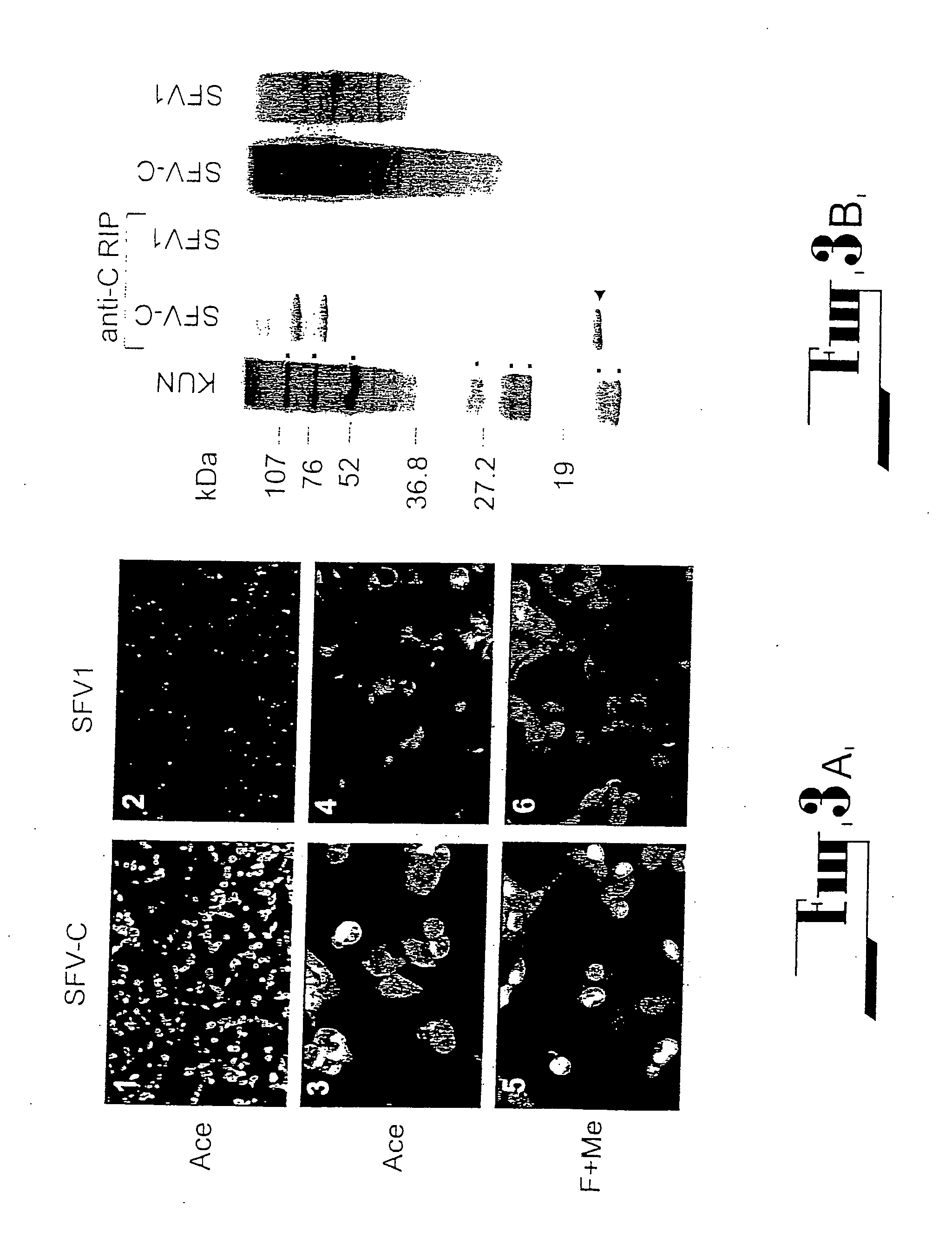
[0100] (i) C20Rep
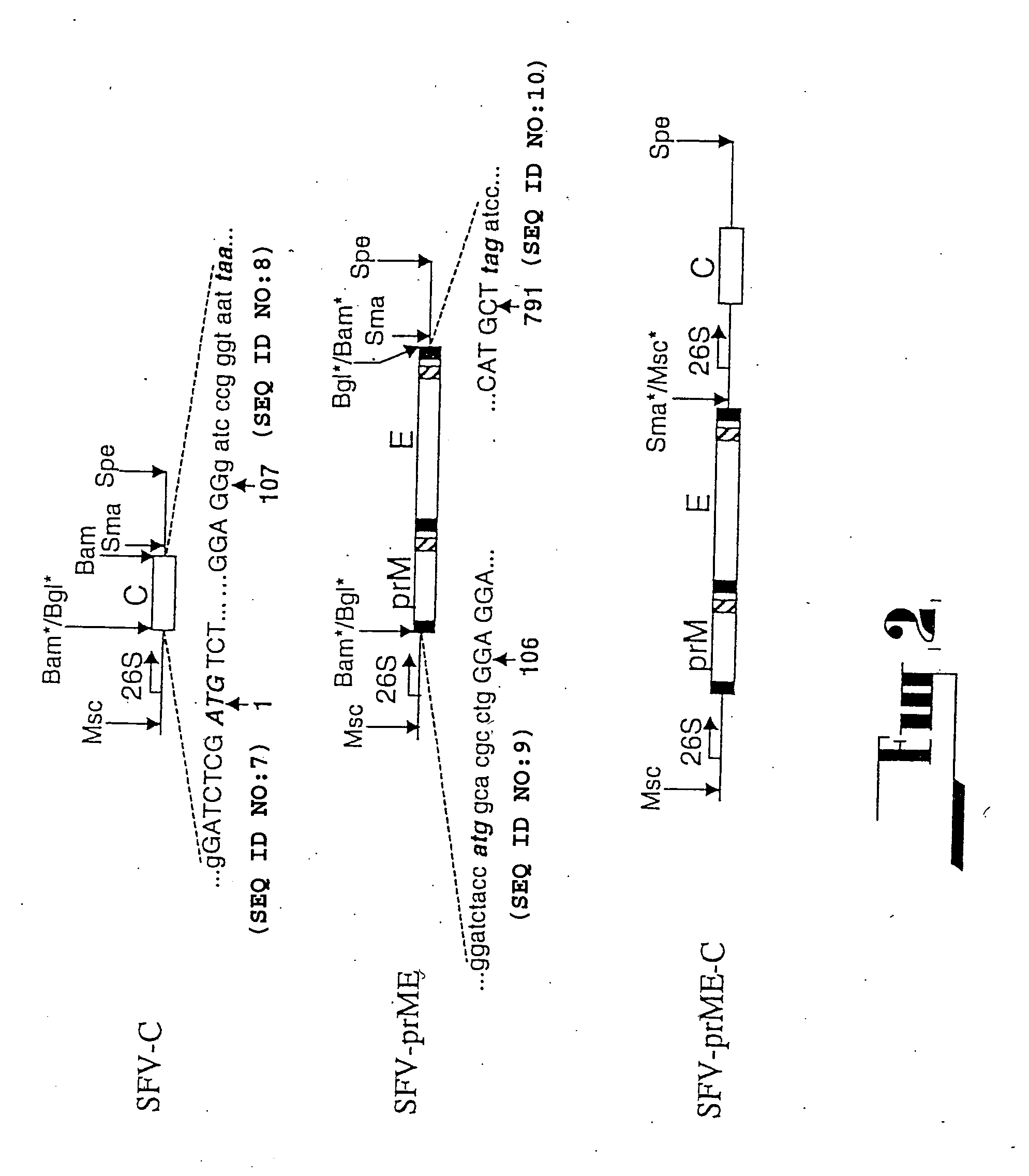
[0101] All deletion constructs were prepared from the cDNA clones used in the construction of the plasmid pAKUN for generation of the infectious KUN RNA (Khromykh and Westaway, J.Virol., 1994, 68:4580-4588) by PCR-directed mutagenesis using appropriate primers and conventional cloning. dME cDNA and its derivatives were deleted from nucleotides 417 to 2404, which represent loss of the signal sequence at the carboxy terminus of C now reduced to 107 amino acids, deletion of prM and E, with the open reading frame resumed at codon 479 in E, preceding the signal sequence for NS1. C20 rep and C2rep cDNAs represent progressive in frame deletions in coding sequence of C leaving only 20 or 2 amino acids of C, respectively, with the open reading frame continued at codon 479 in E, as in dME.
[0102] (ii) FLSDX
[0103] All RT reactions were performed with Superscript II RNase H-reverse transcriptase (Gibco...
example 2
Preparation of Encapsidated Particles and Determination of Their Titer.
[0128] For all infections with encapsidated particles, cell culture fluid was filtered through a 0.45 μm filter (Sartorius AG, Gottingen, Germany) and treated with RNase A (20μg per ml) for 0.5h at room temperature (followed by 1.5h incubation at 37° C. during infection of cells). To prepare partially purified particles, filtered and RNase A treated culture fluids from transfected cells were clarified by centrifugation at 16,000 g in the microcentrifuge for 15 min at 4° C., and the particles were pelleted from the resulting supernatant fluid by ultracentrifugation at 40,000 rpm for 2h at 4° C. in the AH650 rotor of the Sorvall OTD55B centrifuge. The pellets were resuspended in 50 μl PBS supplemented with RNAse A (20μg per ml), left to dissolve overnight at 4° C., and then used for infection of BHK21 cells or for RT-PCR analysis. To determine the titer of encapsidated particles, BHK21 cells on 1.3 cm2 coverslips ...
example 3
Construction of Modified KUN Replicon Vectors and Expression of Heterologous Genes.
Cells.
[0139] BHK21 cells were grown in Dulbecco's modification of minimal essential medium (DMEM, Gibco BRL) supplemented with 10% of fetal bovine serum (FBS). Vero cells were grown in M199 medium (Gibco BRL) supplemented with 5% FBS.
Construction of the Plasmids.
[0140] (I) C20DXrepNeo.
[0141] The dicistronic replicon construct C20DXrepNeo used for generation of replicon-expressing BHK cells (repBHK) was prepared from C20DXrep by cloning an Ires-Neo cassette into the 3′ UTR 25 nucleotides downstream of the polyprotein termination codon. An XmaI-XhoI fragment from ΔME / 76Neo plasmid (Khromykh and Westaway, J.Virol.1997, 71:1497-1505) representing nucleotides 10260 -10422 of KUN sequence, followed by the IRES-Neo cassette and the last 522 nucleotides of KUN sequence was used to substitute XmaI10260-XhoI11021 fragment in C20DXrep construct. Note, that IRES-Neo cassette was initially derived from the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volumes | aaaaa | aaaaa |
| volumes | aaaaa | aaaaa |
| volumes | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com