Reagent sets and gene signatures for renal tubule injury
a technology of gene signatures and reagent sets, applied in the field of reagent sets and gene signatures for renal tubule injury, can solve the problems of renal failure among hospitalized patients, necrotic lesions of the tubules, expensive and long studies, etc., and achieve the effect of increasing the average performance of a depleted s
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of Chemogenomic Reference Database (DrugMatrix™)
[0110] This example illustrates the construction of a large multivariate chemogenomic dataset based on DNA microarray analysis of rat tissues from over 580 different in vivo compound treatments. This dataset was used to generate RTI signatures comprising genes and weights which subsequently were used to generate a necessary set of highly responsive genes that may be incorporated into high throughput diagnostic devices as described in Examples 2-7.
[0111] The detailed description of the construction of this chemogenomic dataset is described in Examples 1 and 2 of Published U.S. Pat. Appl. No. 2005 / 0060102 A1, published Mar. 17, 2005, which is hereby incorporated by reference for all purposes. Briefly, in vivo short-term repeat dose rat studies were conducted on over 580 test compounds, including marketed and withdrawn drugs, environmental and industrial toxicants, and standard biochemical reagents. Rats (three per group) w...
example 2
Preparation of a Chemogenomic Dataset for Late-Onset Renal Tubule Injury
[0115] This example describes methods used to prepare a chemogenomic dataset (i.e., a positive training set) for use deriving a signature for renal tubule injury (i.e., late-onset nephrotoxicity).
[0116] Overview
[0117] 28-day repeat dose studies were conducted on known nephrotoxicants. Doses were chosen that would not cause histological or clinical evidence of renal tubular injury after 5 days of dosing, but would cause histological evidence of tubular injury after 28 days of dosing. Animals were assigned to groups such that mean body weights were within 10% of the mean vehicle control group. Test compounds were administered either orally (10 ml of corn oil / kg body weight) or by intra-peritoneal injection (5 ml of saline / kg body weight). Animals were dosed once daily starting on day 0, and necropsied 24 hrs after the last dose following an overnight fast on day 5 (n=5) and day 28 (n=10). An equivalent number o...
example 3
Derivation of a Predictive Renal Tubule Injury Signature
[0138] Overview
[0139] The support vector machine algorithm was trained to classify experimentally confirmed nephrotoxicants from non-nephrotoxicants using the data acquired in Examples 1 and 2 above. A linear classifier (i.e., gene signature) was derived using kidney expression profiles from rats treated with 15 nephrotoxicants that induce renal tubular injury after 4 weeks of daily dosing, and 49 non-nephrotoxicants known not to induce renal tubular injury under subchronic dosing conditions.
[0140] Gene Signature Derivation
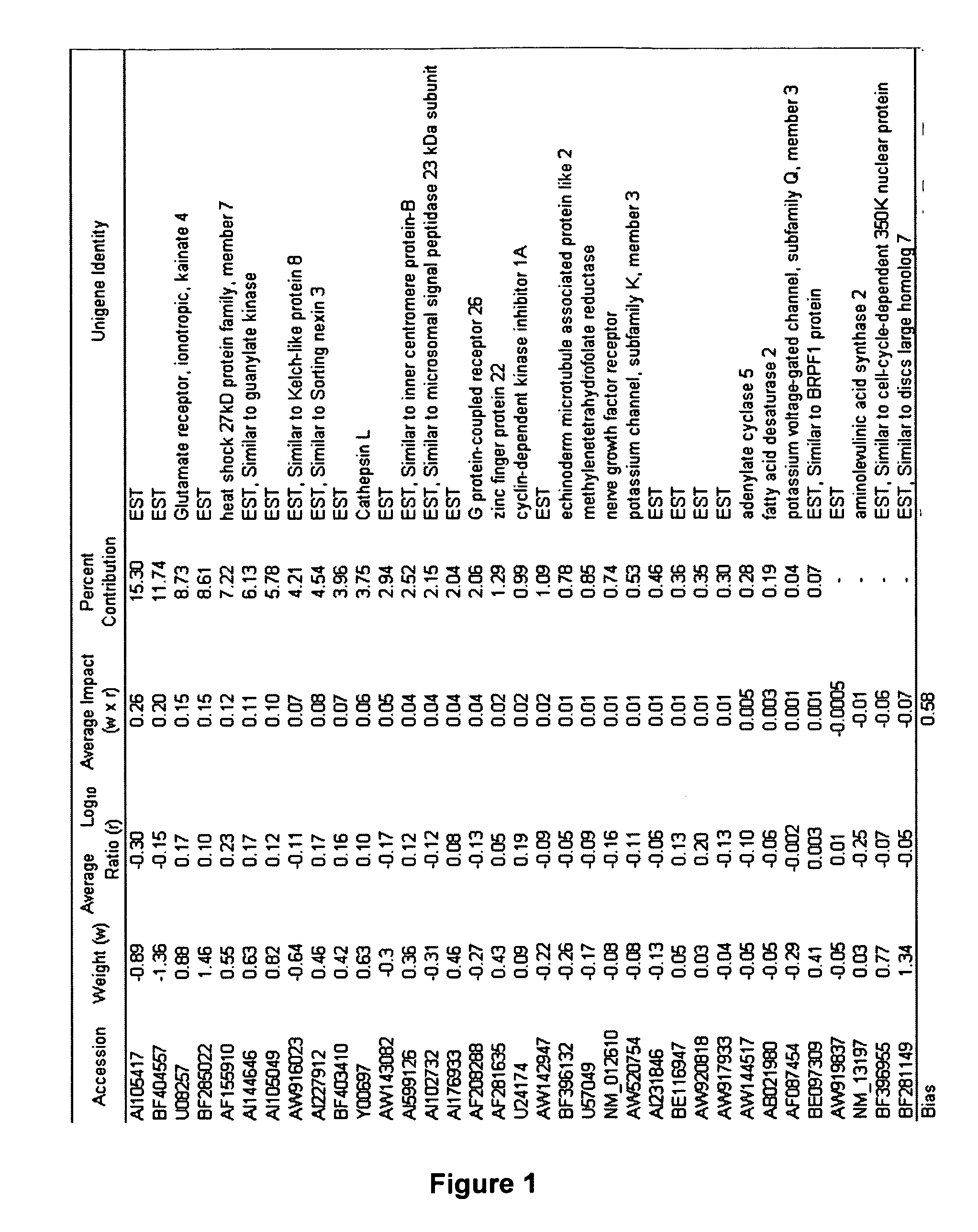
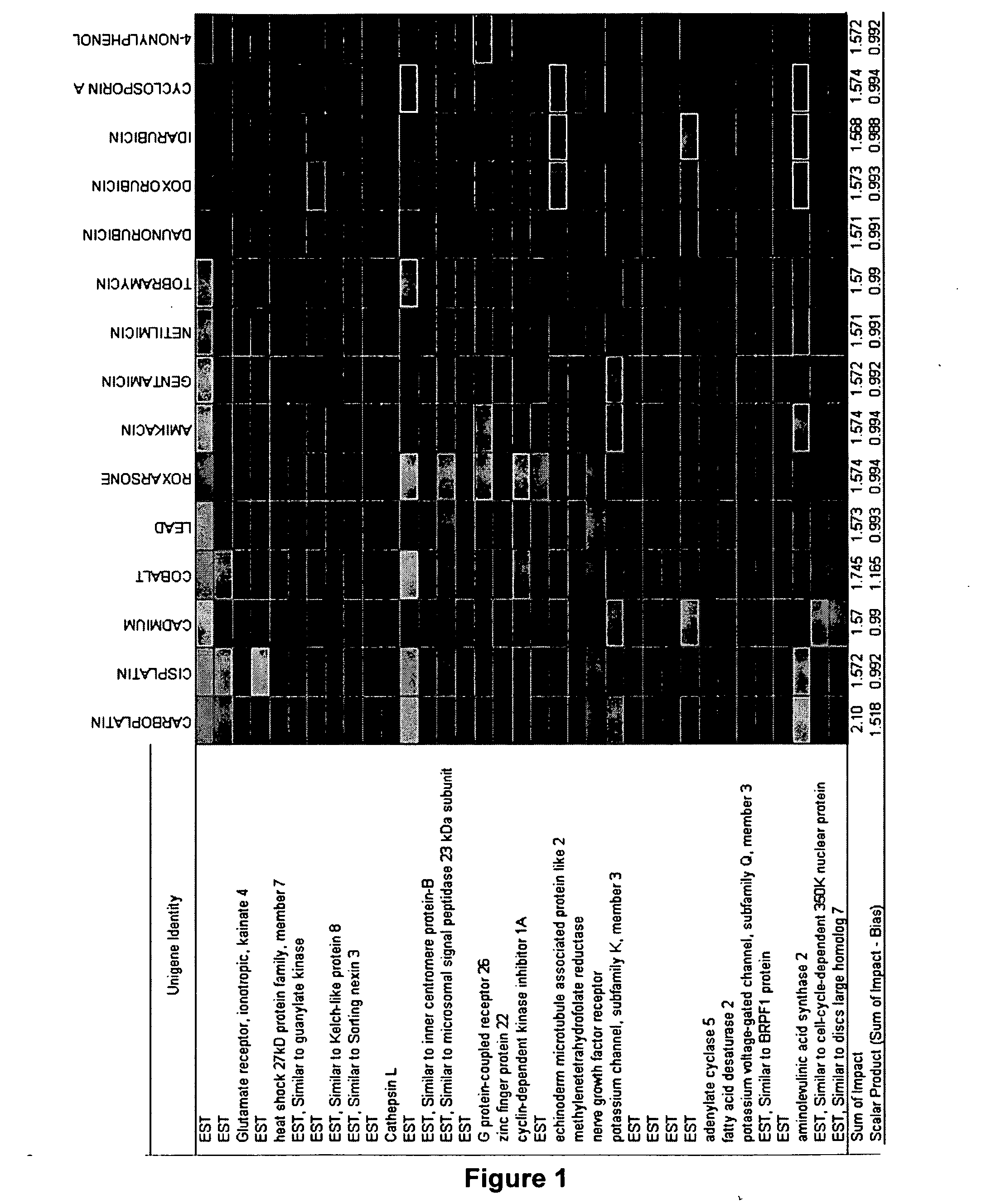
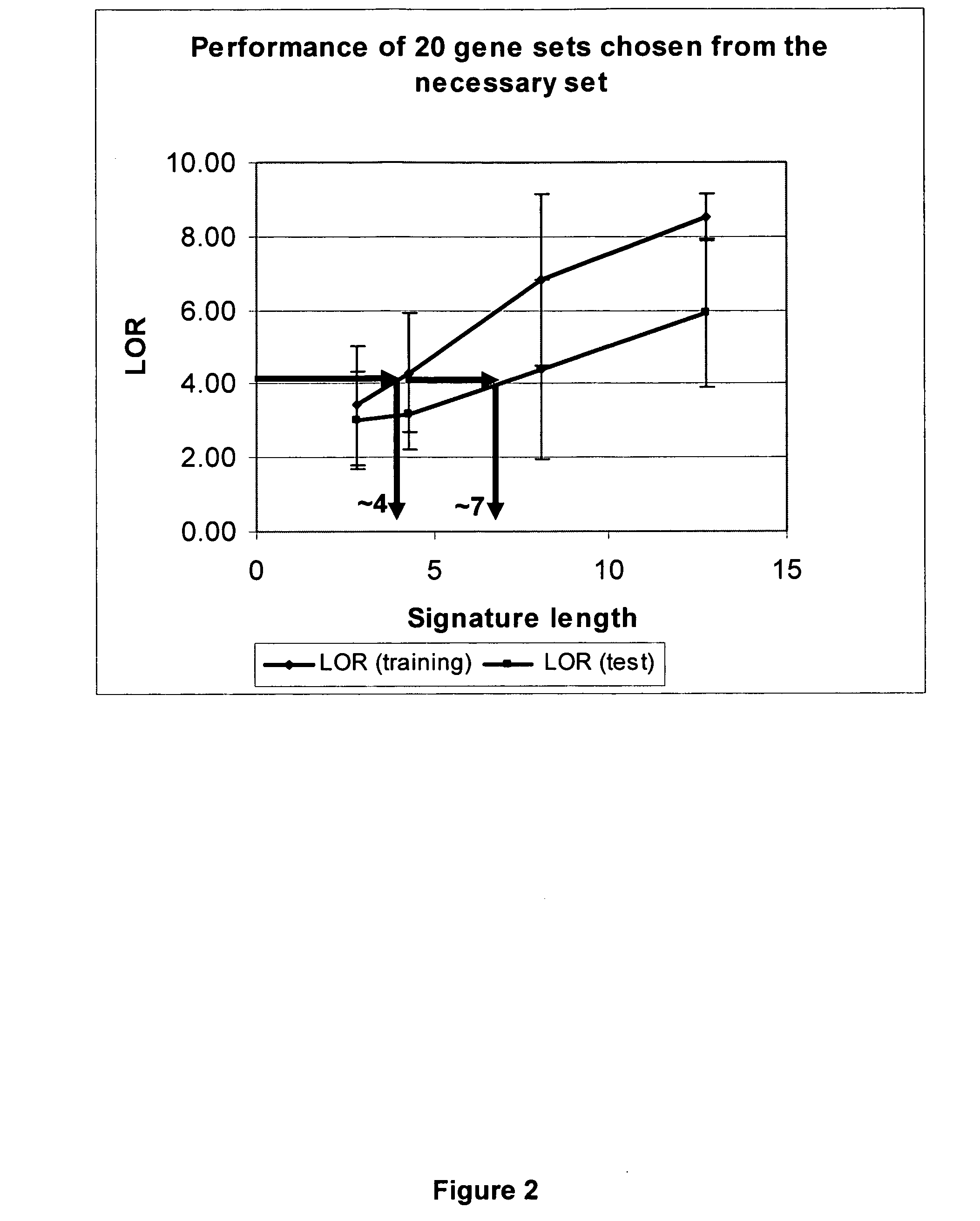
[0141] To derive the gene signature, a three-step process of data reduction, signature generation and cross-validation of the predictive signature was used. A total of 7478 gene probes from the total of 10,000 on the CodeLink™ RU1 microarray were pre-selected based on having less than 5% missing values (e.g., invalid measurement or below signal threshold) in either the positive or negative class of the tr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com