Direct differentiation of cardiomyocytes from human embryonic stem cells
a technology of embryonic stem cells and cardiomyocytes, which is applied in the field of stem cell differentiation induction, can solve the problems of chronic heart failure, loss of functional cardiomyocytes, and insufficient replacement of injured or dead cardiomyocytes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Characterization of END2 Conditioned Media
I. Materials and Methods
(i) Preparation of END2 conditioned medium
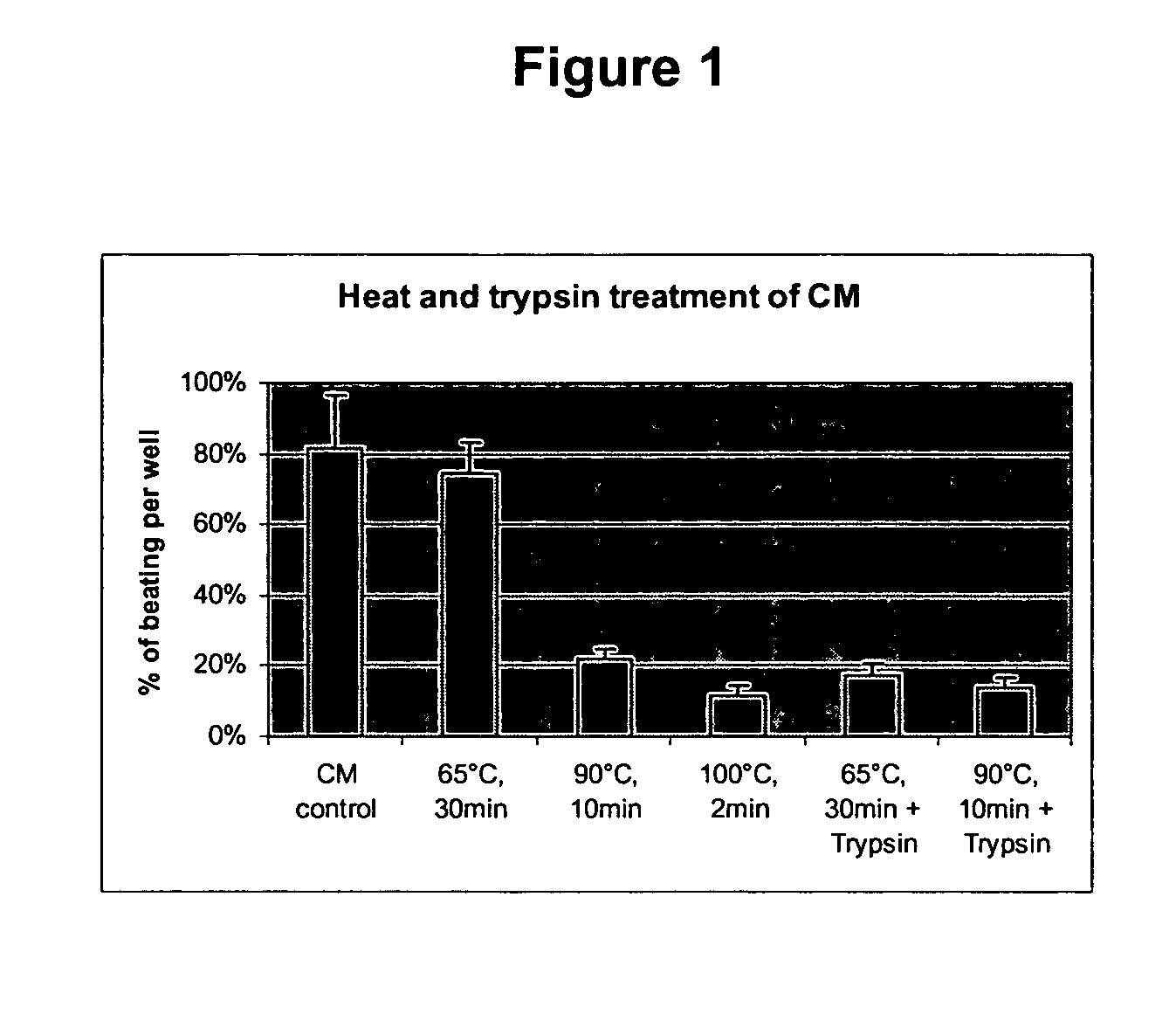
[0123] END2 cells secrete a factor into culture medium that can induce cardiomyocyte differentiation in hES cells. END2 conditioned media (END-CM) is defined as serum-free (SF) media that has been used to culture END-2 cells for four days or more and contains cardiomyocyte-inducing factor / s and / or bioactivity thereafter. hES cells are Human Embryonic Stem Cells. END2 cells are a visceral-endoderm-like cell line derived from P19, a mouse embryonal carcinoma cell line.
[0124] For the production of END2-CM END2 cells were seeded at a density of 1.4×105 cells / cm2 in a T175 flask (Nunc) and were grown for about 3 days in Dulbecco's Modified Eagle's Medium (DMEM) / F12 media (Invitrogen) supplemented with 7.5% fetal calf serum (FCS; Hyclone) until confluency. The confluent END2 cell layer was washed once in PBS+ (Gibco) and serum free (SF) medium was used for conditioning. SF med...
example 2
Microarray Data Analysis to Identify Cardiomyocyte Inducing Factors Produced by END2 Cells
I. Materials and Methods
(i) Microarray Analysis
[0162] The Microarray experiments were conducted using Agilent Mouse 20k Developmental Oligo array or genome 40 k Oligo array technology. MES-1 was used as a reference cell line as it derived from the same parental cell p19 EC cell line as END2 but negative in its induction of cardiac differentiation. Two replica array datasets with dye swap were generated in two independent array experiments. Only features' expression calls were positive and significant above background assigned by the “Agilent G2565AA Feature Extraction Software” were accepted for analysis. The array data files were imported into GeneSpring 7.1 (Silicon Genetics) for data analysis. The expression ratios (Cy5 / Cy3: ratio of the median) were calculated and transformed to log2. Log2 expression ratios were then normalized by intensity-dependent LOWESS normalization. The different...
example 3
Prostaglandin I2 (PGI2) is an Active Component of END2 CM and Able to Increase hES Cell Cardiac Differentiation
I. Materials and Methods
(i) Medium Supplementation with PGI2
[0173] Prostaglandin I2 or PGI2-Na {(5Z, 9α, 11α, 13E, 15S)-6,9-Epoxy-11,15-dihydroxyprosta-5,13-dien-1-oic acid Sodium salt] was obtained from Sigma-Alorich. It was dissolved in 90% Ethanol at 2 mM as stock solution and then diluted with PBS before adding to bSFS at final concentration of 0.2 uM, and 2 uM. Medium change with addition of respective amounts of PGI2 was performed at day 3, 6 and 9 after EB formation.
II. Results
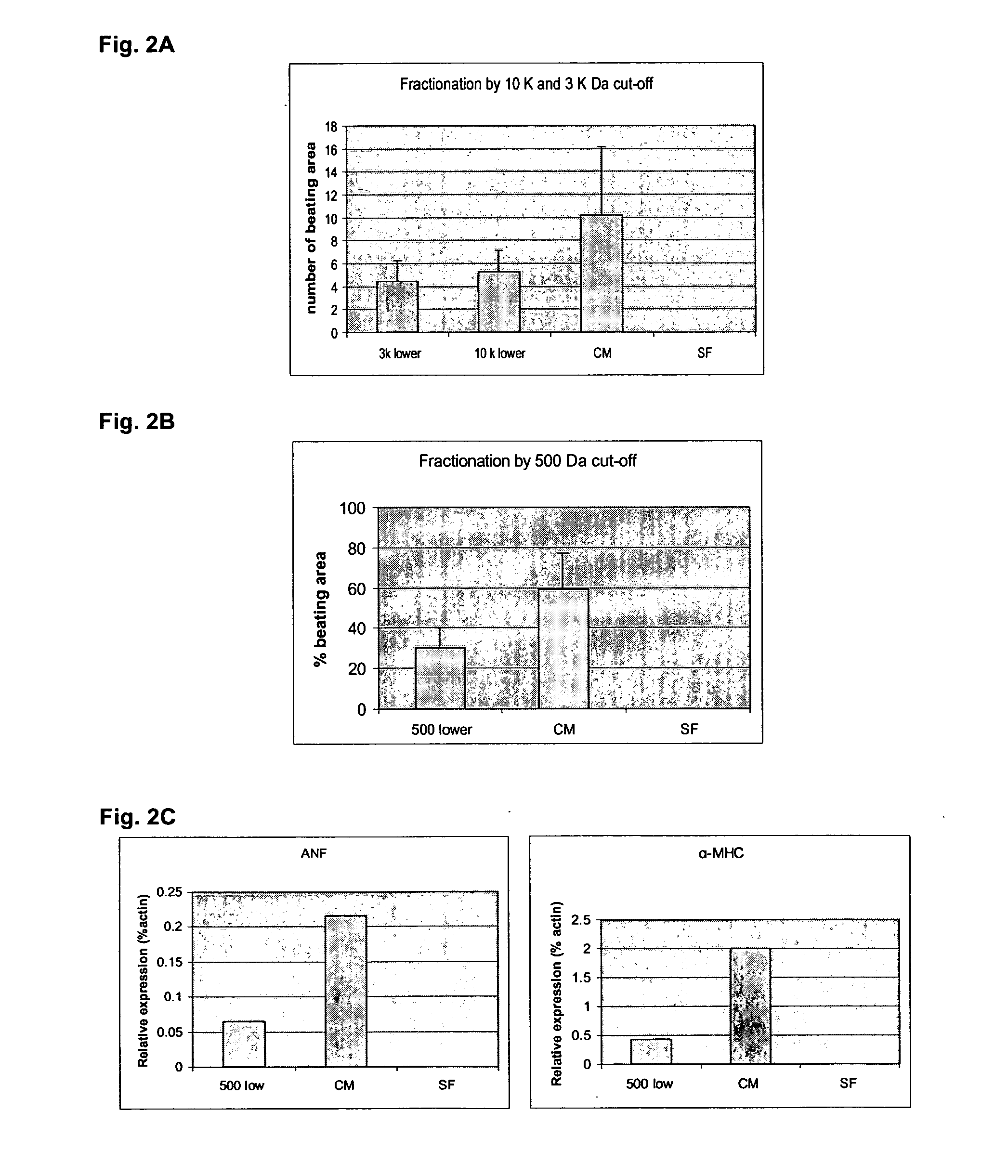
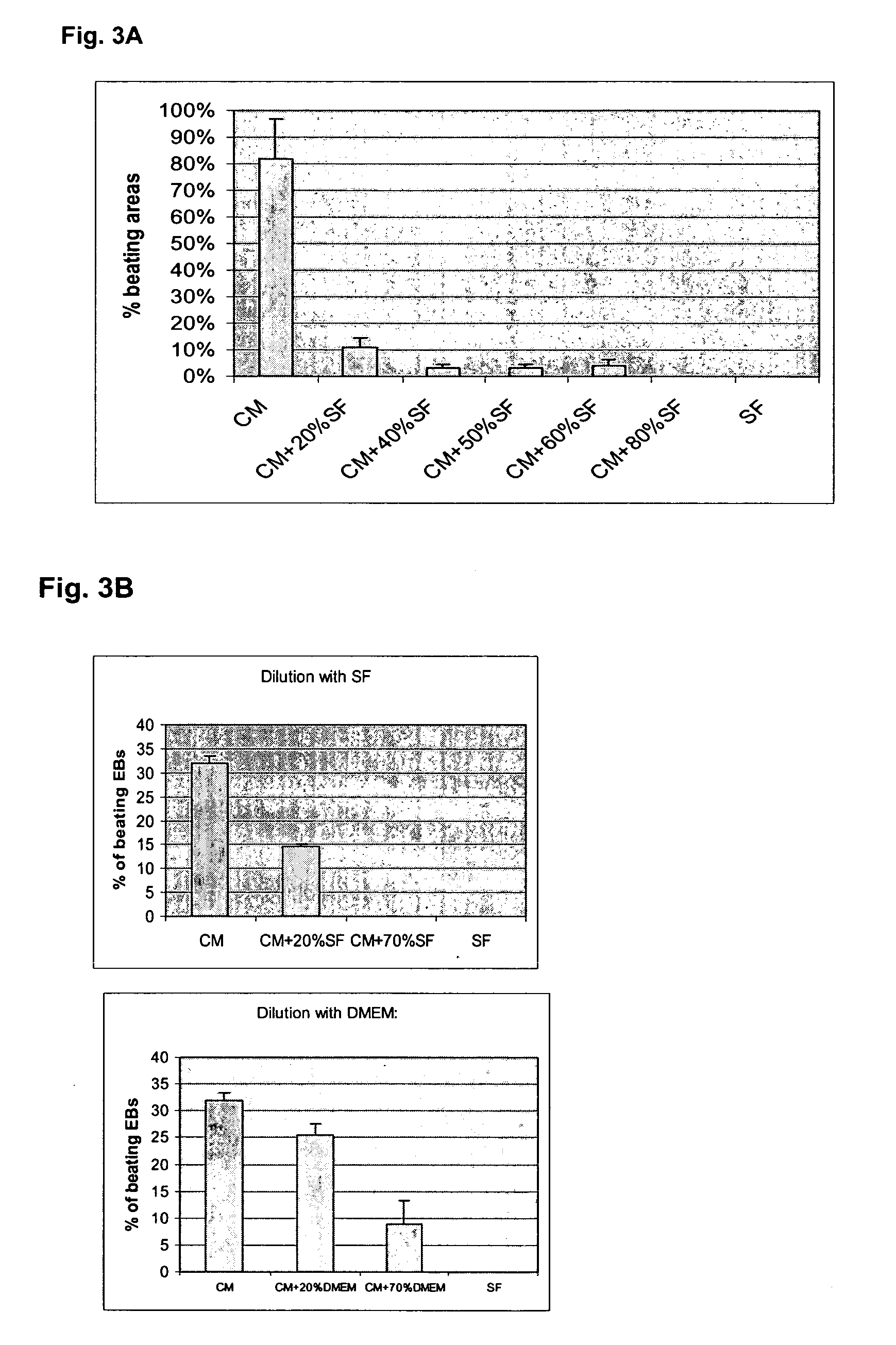
[0174] Factors found in the END2 conditioned media (CM) and involved in the ability of END2 cells to induce hES cell cardiac differentiation were identified. Accumulated data from studies on CM (physiochemical properties and size fractionation) from the previous examples suggested that the inducing factor in END2-CM is potentially a small molecule that is not a protein. Microarray analy...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com