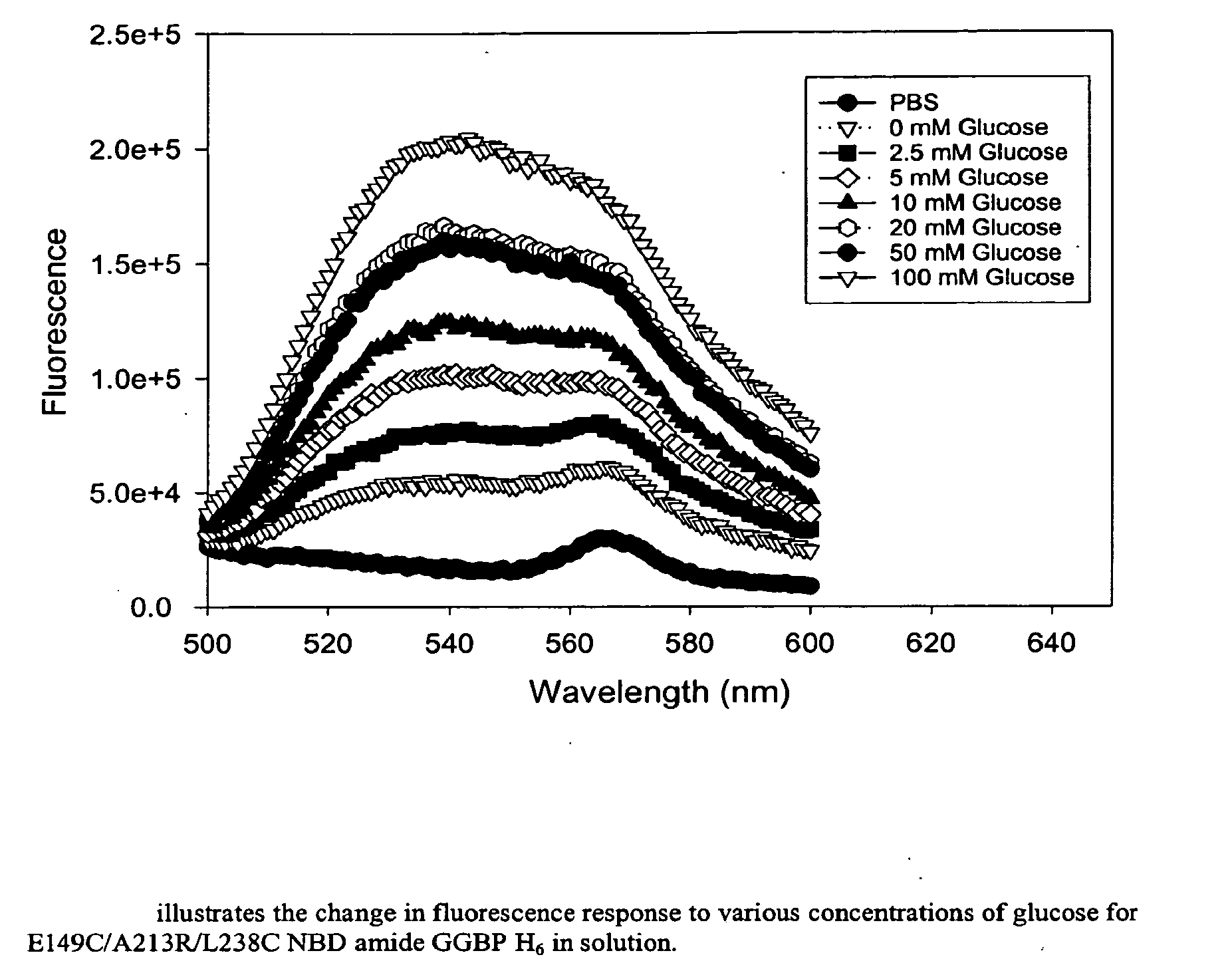
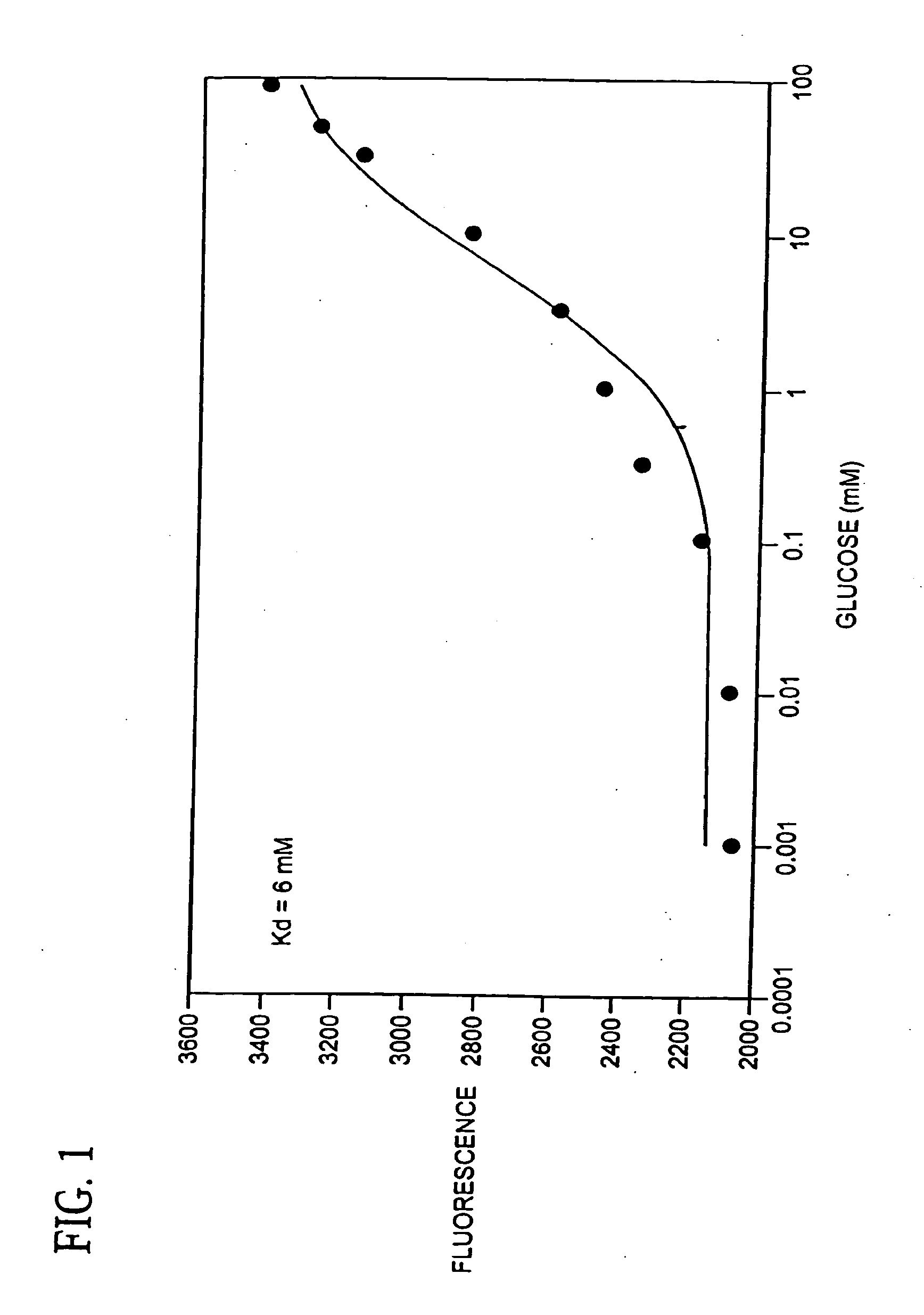
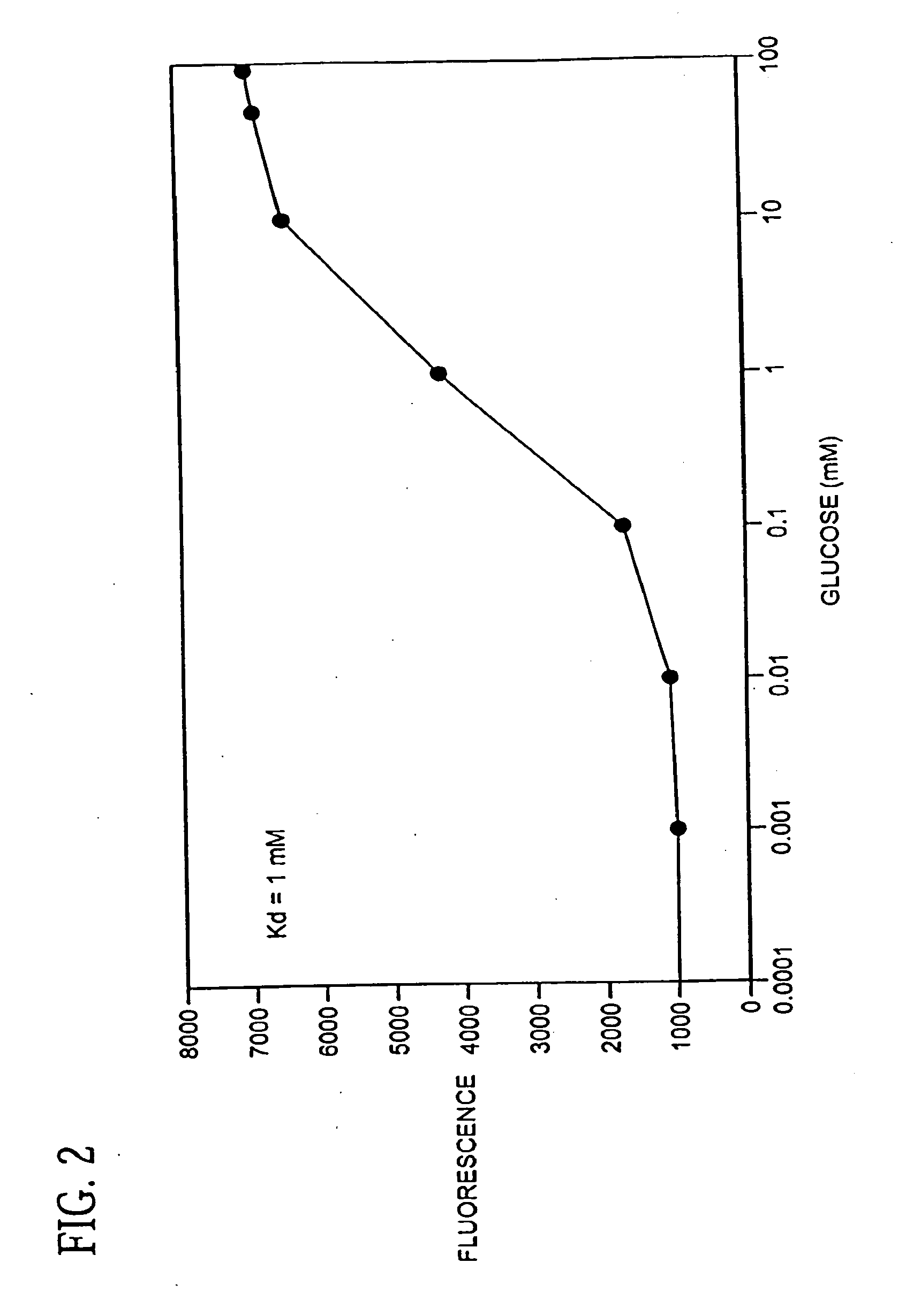
Binding proteins as biosensors
a technology of binding proteins and biosensors, applied in the field of biotechnology, can solve problems such as the inability the inability of protein and the associated reporter group to bind diagnostically important sugars, and the difficulty of patient compliance,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Method for the Expression and Purification of Mutant Proteins without Histidine Tags
[0050] GGBP is coded by the Mg1B-1 gene in E. coli. This protein was altered by introducing the amino acid cysteine at various positions through site-directed mutagenesis of the Mg1B-1 gene. These proteins were then expressed in E. coli and purified.
[0051] Cassette mutagenesis of Mg1B-1 was accomplished as follows. The wild-type Mg1B-1 gene was cloned into a pTZ18R vector (Dr. Anthony Cass, Imperial College, London, England). Mutant plasmids were generated from this parent plasmid using cassette mutagenesis producing randomized amino acid sequences, essentially as described by Kunkel (1991) and cloned in E. coli JM109 (Promega Life Science, Madison, Wis.). Mutant plasmids were identified by sequencing. The mutant protein was induced in JM109 and purified as described below. An E. coli JM109 colony containing the mutant plasmid was grown overnight at 37° C. with shaking (220 rpm) in LB broth contain...
example 2
Expression and Purification of Mutant GGBPs Containing Histidine Tags
[0053] GGBP mutants were engineered by either site-directed mutagenesis or cassette mutagenesis. Site-directed mutagenesis (QuikChange, Stratagene, La Jolla, Calif.) was performed to alter individual amino acids in the pQE70 vector by replacing one amino acid with another, specifically chosen amino acid. The cassette mutagenesis method (Kunkel 1991) was performed to randomize amino acids in a specified region of the GGBP gene. The mutated cassettes were then subcloned into the pQE70 expression vector.
[0054] The pGGBP-His plasmid contained the GGBP gene cloned into the pQE70 expression vector (Qiagen, Valencia, Calif.). This construct places six histidine residues on the C-terminus of the GGBP gene. E. coli strain SG13009 was used to overexpress mutant GGBP-His following standard procedures (Qiagen). After overexpression of a 250 mL culture, the cells were collected by centrifugation (6000 rpm) and resuspended in ...
example 3
Labeling of Mutant GGBPs
[0055] An aliquot of mutant GGBP containing cysteine (4.0 nmol) in PBS was treated with 2 mM dithiothreitol (5 μL, 10 nmol) for 30 min. A stock solution of N,N′-dimethyl-N-(iodoacetyl)-N′-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)ethylenediamine (IANBD amide, 0.5 mg) was prepared in DMSO (100 μL, 11.9 mM) and 3.36 μL (40 nmol) was added to the protein. The reaction proceeded at room temperature for 4 h on a Dynal rotamix in the dark. The labeled protein was purified by gel filtration on a NAP-5 column (Amersham Pharmacia). The labeling ratios were determined using an estimated extinction coefficient (50 mM−1 cm−1) for GGBP that was calculated in GeneWorks 2.45 (IntelliGenetics), ε478 (IANBD amide)=25 mM−1 cm−1), and a measurement of O.D. for a standard solution of IANBD amide at 280 nm and 478 nm. The dye concentration in the protein was calculated as Cdye=A478 / ε478. The absorbance of protein at 280 nm was calculated as Aprot(280)=Atotal(280)−Adye(280), where Adye(...
PUM
| Property | Measurement | Unit |
|---|---|---|
| excitation wavelength | aaaaa | aaaaa |
| emission wavelength | aaaaa | aaaaa |
| excitation or emission wavelengths | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com