Amino acid substituted molecules
a technology of amino acid and substituted molecules, applied in the field of amino acid substituted molecules, can solve the problems of largely ineffective and inefficient modification of molecules, including proteins, presently
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Prophetic
Design of Site-Specific PEGylated Proteins
[0465] The design of a pegylated GM-CSF, Erythropoietin (EPO), Human Growth Hormone, Phenylalanine hydroxylase, urikase, Factor VII, follitropin, G-CSF, or other target molecule may comprise a multi-step process. In the case of EPO, which wild type sequence contains two methionine amino acids—including one at the amino terminus, only one methionine would require substitution. In the case of G-CSF, the wild type sequence does not contain any arginine residues. Thus, an arginine residue could be introduced at any desirable location in the molecule and subsequently substituted or replaced with a non-natural amino acid. Likewise, for Human Growth Hormone, the wild type sequence only contains a single tryptophan residue, phenylalanine hydroxylase contains only 3 methionine residues and 3 tryptophan residues, and follitropin contains only 5 methionine residues.
[0466] In an optional first step, existing specific target wild type amino a...
example 2
Prophetic
Site-Specific PEGylation of GM-CSF
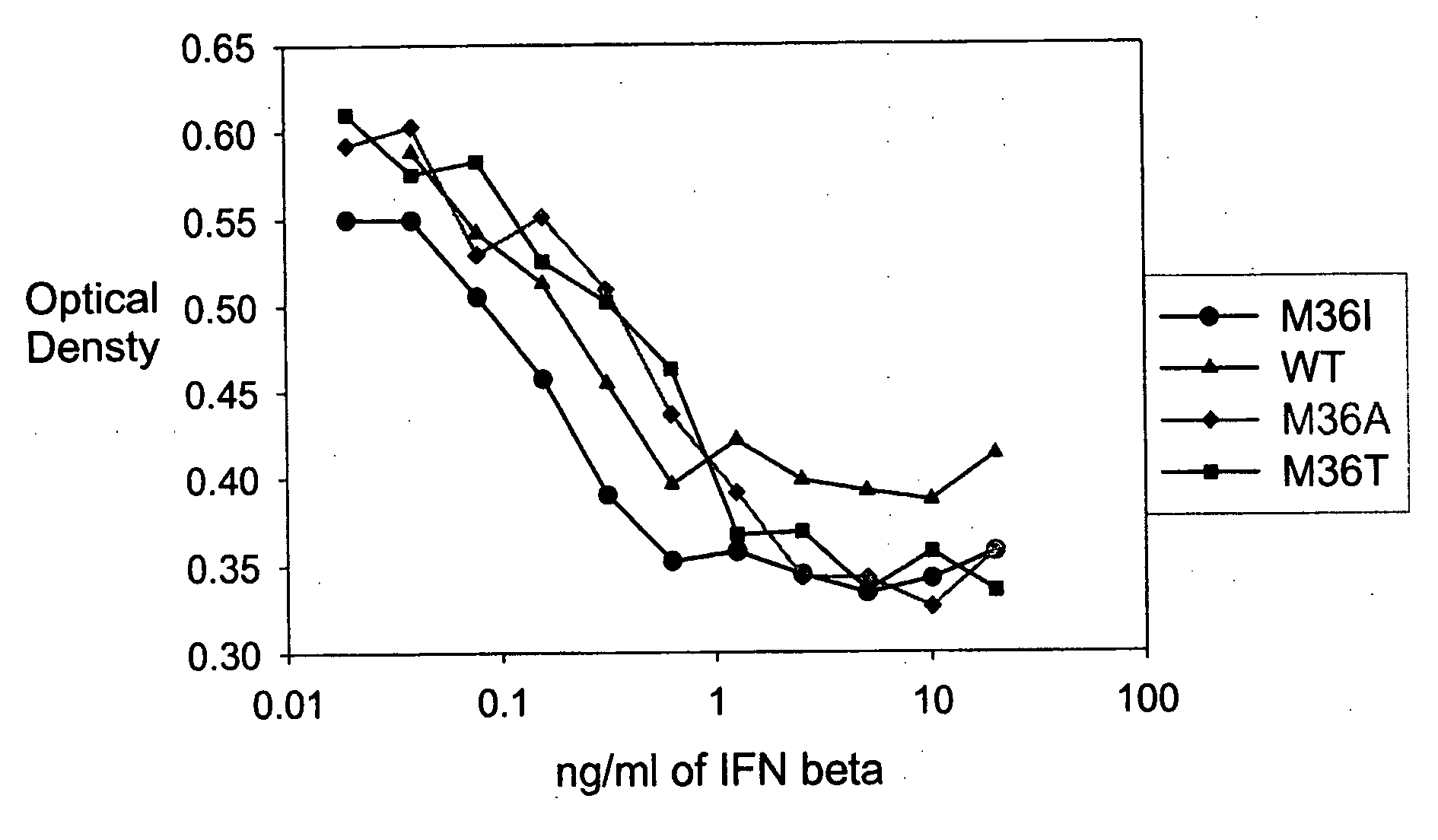
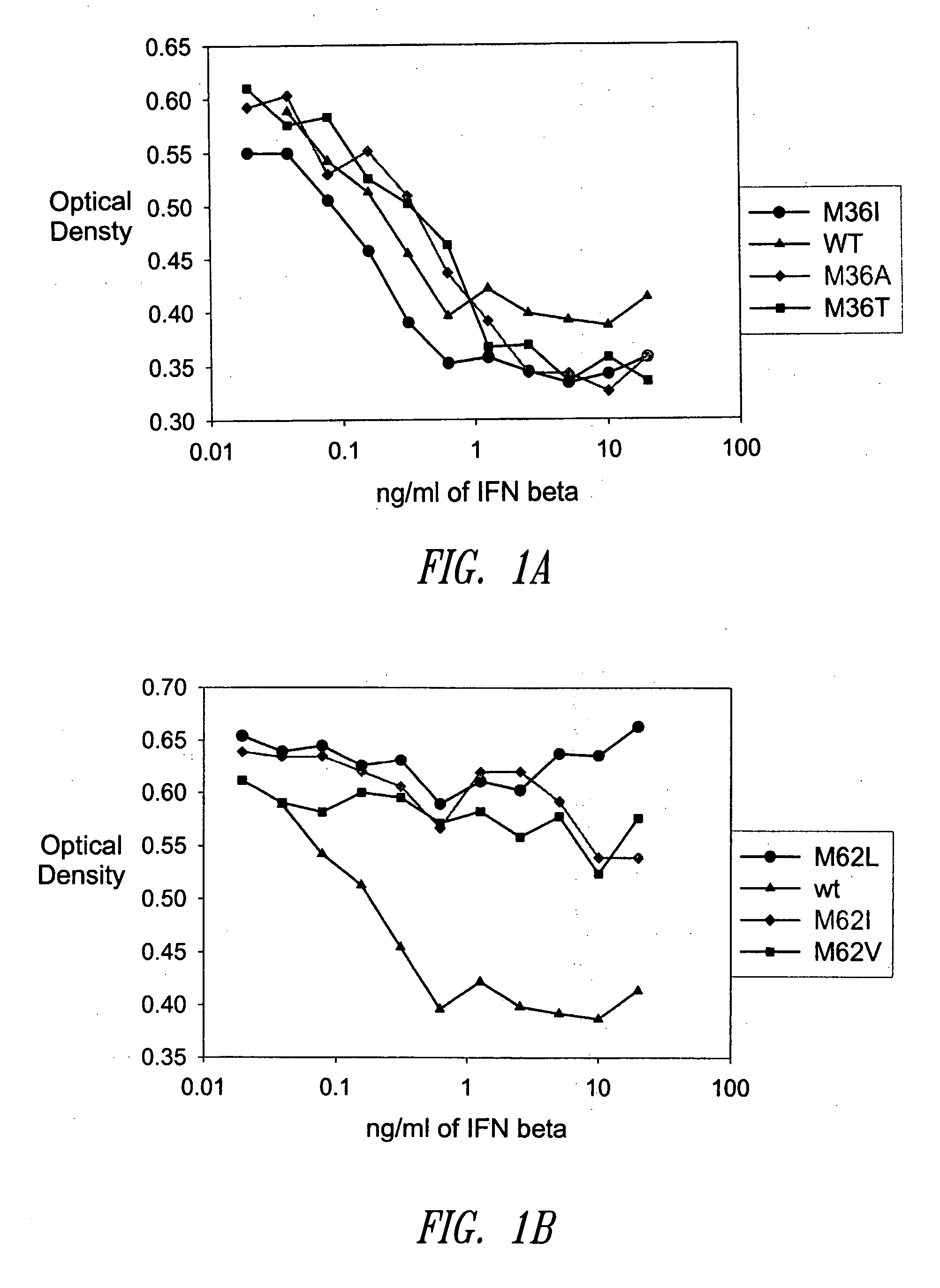
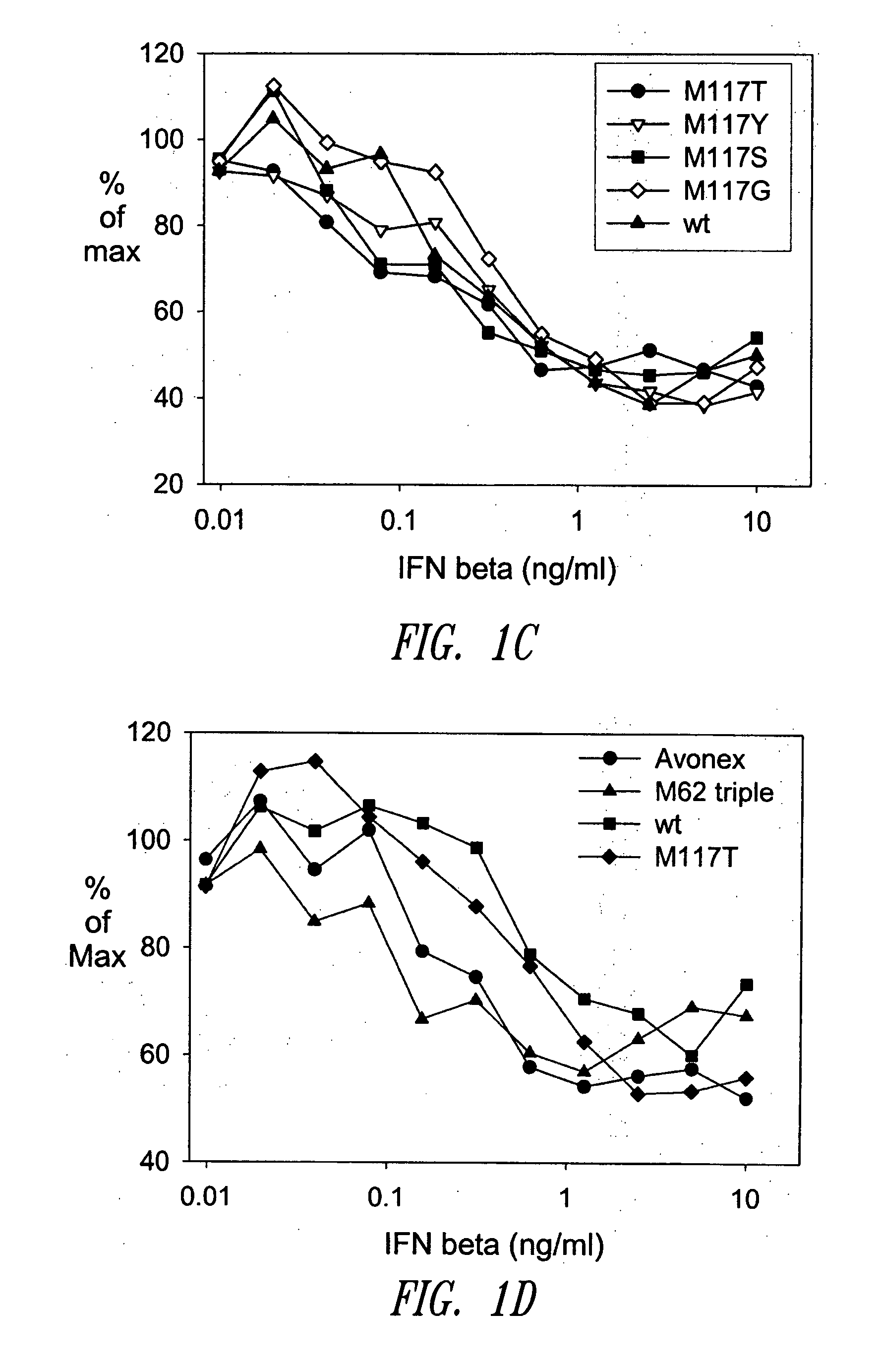
[0471] A GM-CSF molecule contains four wild type methionine amino acid residues at positions 36, 46, 79 and 80. There are at least two possibilities for inserting a site-specific methionine analog into GM-CSF for use as an anchoring residue for pegylation.
[0472] One option would be to retain one of the four methionine residues in the GM-CSF molecule and replace the three other methionine residues with other naturally occurring amino acid residues. Selecting which three methionine residues will be replaced and / or selecting which naturally occurring amino acid residues shall replace the three wild type methionine residues may be determined, in part, by evaluating energy calculations as described herein. Additionally, replacement amino acids may be selected by alignment of nucleic acid or amino acid sequences of related genes or proteins, respectively. The sequences may be from the same species or different species.
[0473] A second option w...
example 3
Prophetic
Energy Calculations for Site-Specific PEGylated GM-CSF
[0474] Energy calculations for the target molecule discussed in the previous Example may be conducted by any known method, some of which are described herein. The sequence and number of energy calculations may be performed in a number of ways. For example, a point mutation calculation may be performed for each selected methionine position (which include positions 36, 46, 79 and 80). Alternatively or additionally, combination mutation calculations may be performed for all four methionines such that one methionine is retained in its wild type position, while the other three methionine residues will be varied simultaneously to other naturally occurring amino acids. In this manner, it may be determined whether all four methionine residues will be replaced with other amino acid residues, or if one methionine residue will be retained while the other three are replaced with other naturally occurring amino acid residues.
[0475...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting temperature | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com