Methods for cloning small RNA species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
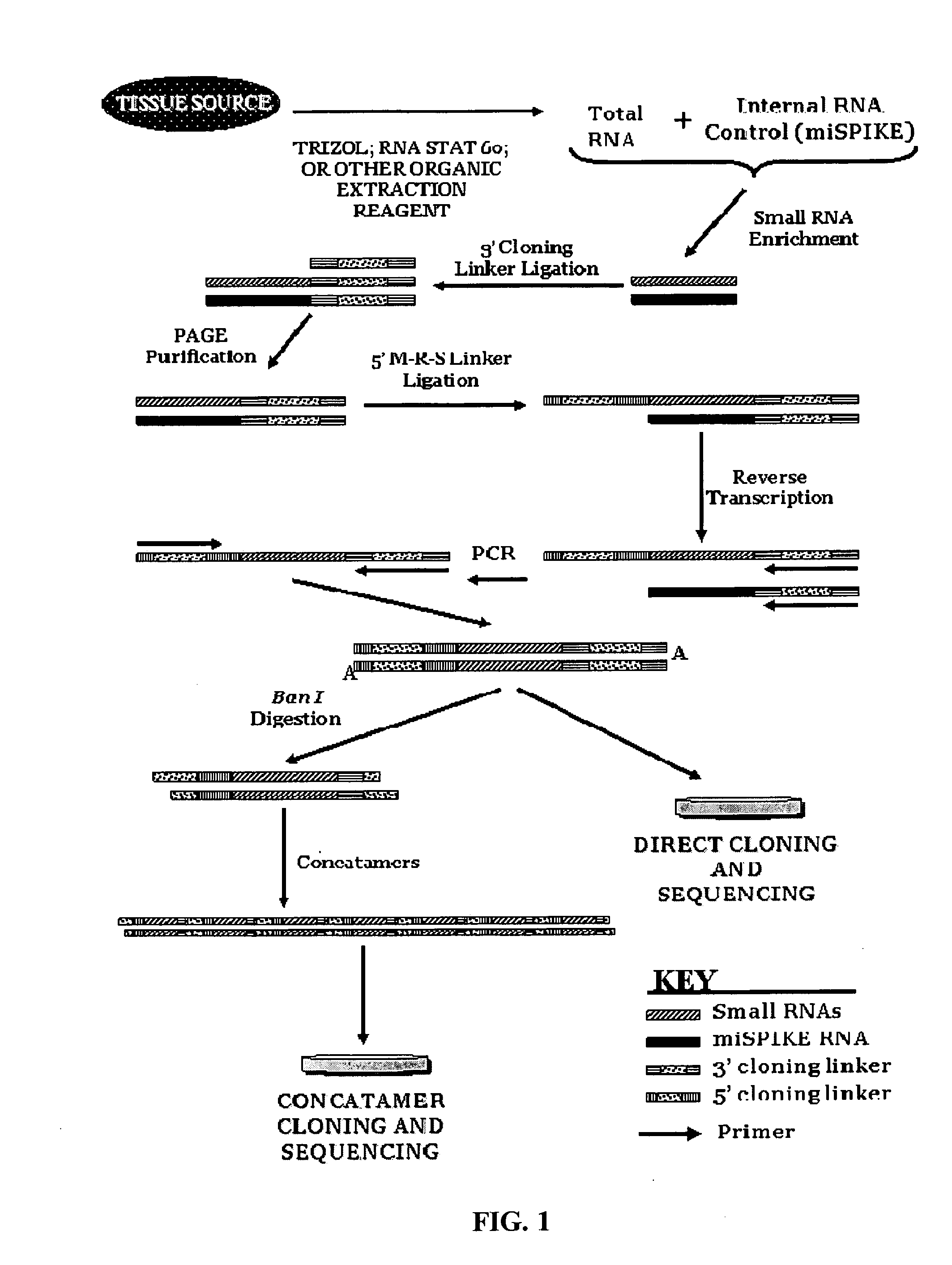
[0049]This example provides a protocol for practicing the proposed microRNA cloning process. Total RNA is prepared using methods well-known in the art, such as the use of the mirVana RNA isolation kit (Ambion®) following the Ambion® protocol. RNA isolation methods utilizing glass fiber filters (GFF) or silicate adsorption should be avoided as these “rapid” methods for RNA isolation often deplete the small RNA pool present in a natural sample. Organic extraction reagents such as Trizol or RNA STAT 60 are preferred. After the RNA is extracted and purified, the sample is enriched for small RNA species. If practical, it is preferred to employ at least 50 μg to 100 μg of total RNAs for small RNA enrichment however lower input mass amounts can be used when sample is limited. The mass of RNAs in the miRNA size range of 18 nt to 26 nt is a very small fraction of the total RNA present, and size selection at this stage of library construction is essential to the quality of the end product. Th...
example 2
[0074]The following example demonstrates the use of the method of the invention to isolate novel miRNAs from tissue.
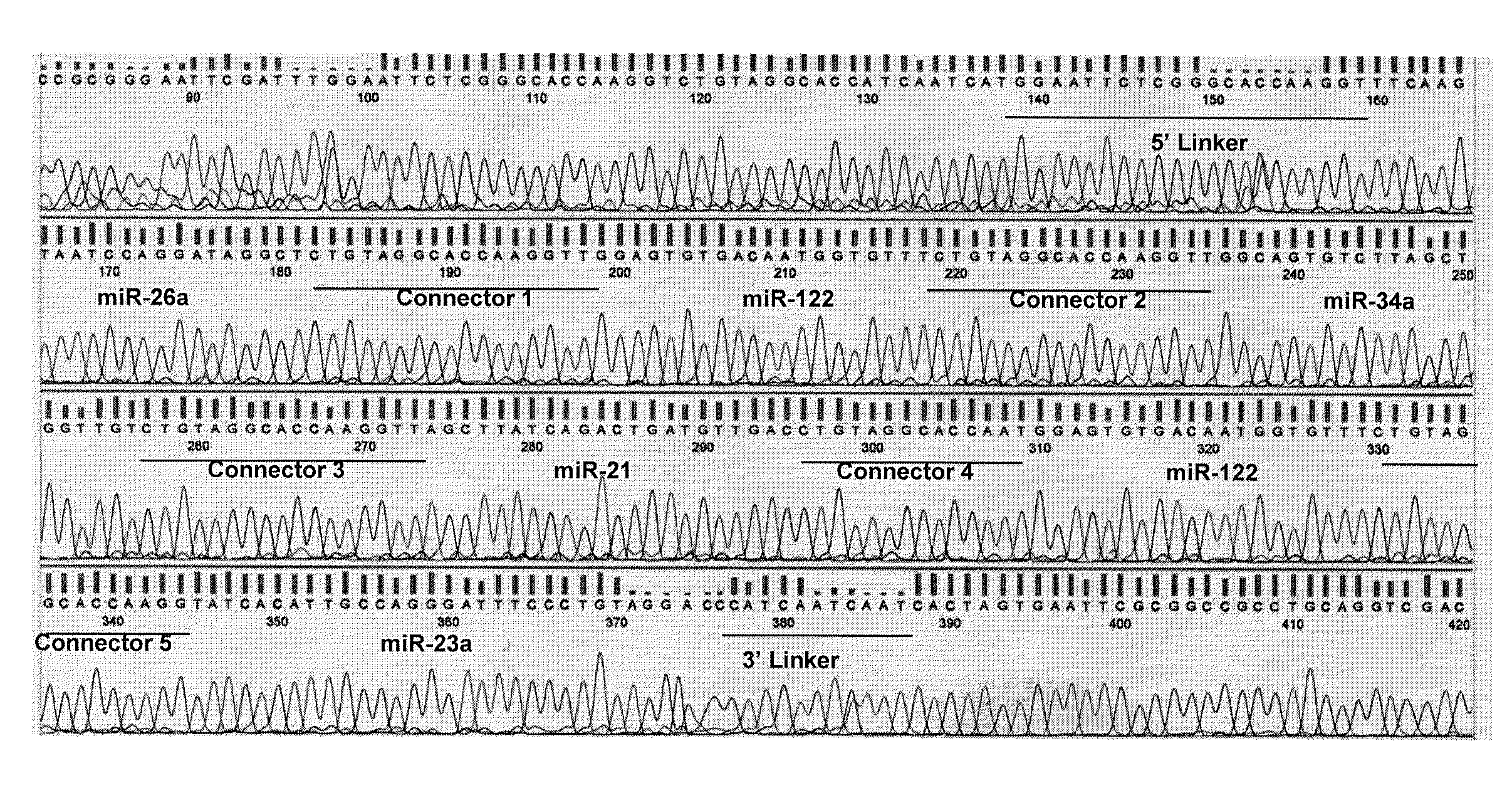
[0075]Total RNA was prepared from brain, heart, lung, liver and kidney tissue of the South American marsupial species Monodelphis domestica using the mirVana RNA isolation kit (Ambion®, Austin, Tex.) according to the manufacturer's protocol. Between 100 μg and 200 μg of total cellular RNA was obtained from each tissue fragment. A 100 μg RNA pool, comprising 20 μg of RNA from each tissue, was used as the RNA source for the experiment. This pooled RNA sample was spiked with 10 pmole of the 21 nt internal control ORN marker “miSPIKE” and size fractionated on a 12% denaturing (7M urea) polyacrylamide gel at 275 volts for 90 minutes.
SEQ ID NO. 15′-rCrUrCrArGrGrArUrGrGrCrGrGrArGrCrGrGrUrCrU-3′
[0076]The gel was stained with GelStar® and RNAs were visualized under UV excitation. A 4 mm square gel slice was excised using the miSPIKE ORN marker as a size guide. The gel slice was...
example 3
[0089]The following example demonstrates the use of the method of the invention to isolate novel piRNAs from tissue.
[0090]Unlike miRNAs which are expressed in all tissues, other classes of small RNAs have limited tissue distribution. The Piwi associated RNAs (piRNAs) are a different class of small RNA which are specific to gonads (ovary and testis). The piRNAs are longer than miRNAs, and usually are 26-32 nt long. The method of the invention was used to isolate and sequence identify novel piRNAs. Total RNA was prepared from testis of the South American marsupial species Monodelphis domestica using the mirVana RNA isolation kit (Ambion®, Austin, Tex.) according to the manufacturer's protocol. Between 100 μg and 200 μg of total cellular RNA was obtained from each tissue fragment. 100 μg testis derived RNA was employed for the experiment. This RNA sample was spiked with 10 pmole of the 31 nt internal control ORN marker “piSPIKE” and size fractionated on a 12% denaturing (7M urea) polya...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Molar density | aaaaa | aaaaa |
| Length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com