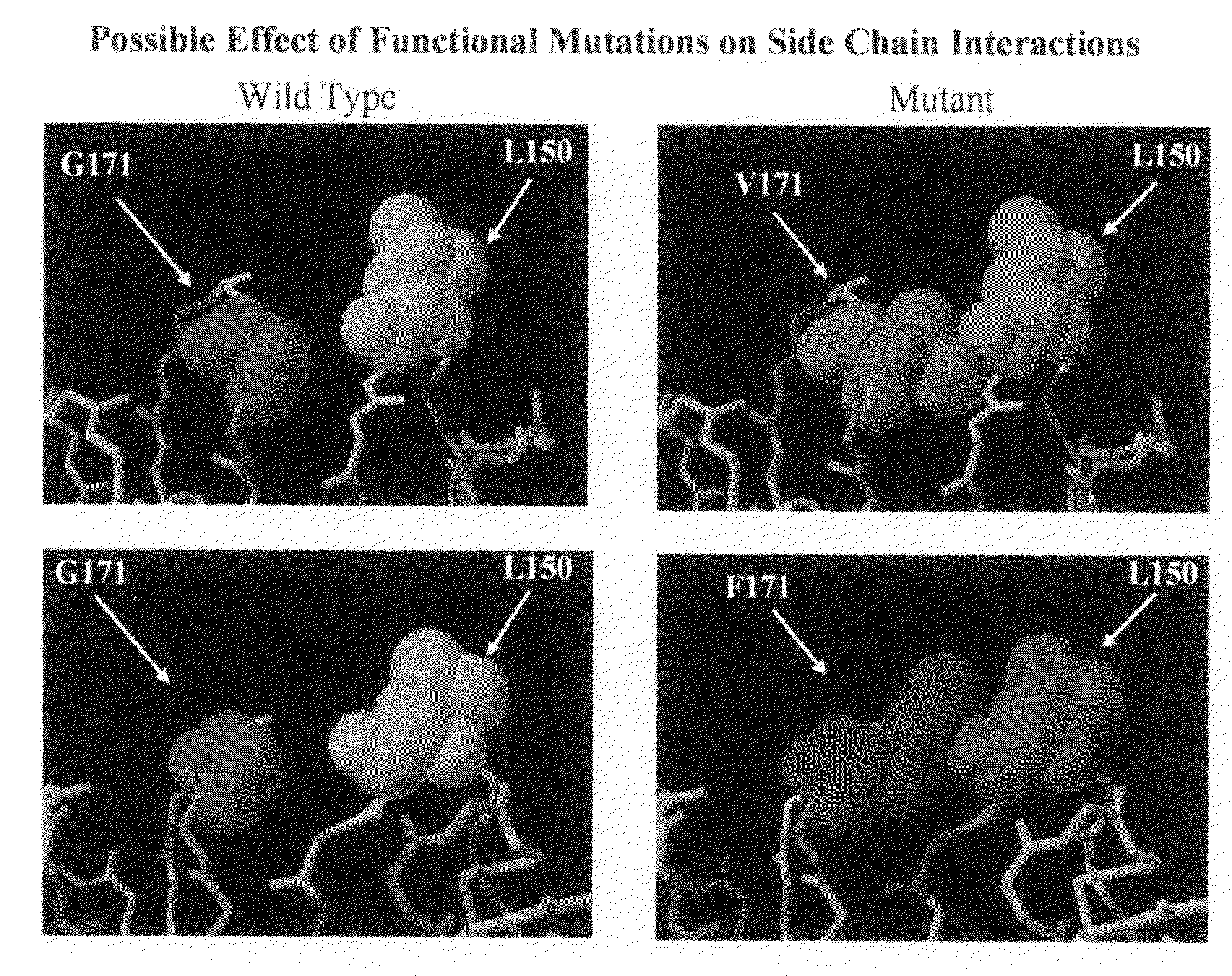
HBM variants that modulate bone mass and lipid levels
a technology of lipid levels and variants, applied in the field of genetics, genomics and molecular biology, can solve the problems of increasing the risk of aging, and reducing the effect of bone resorption and formation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0718]The propositus was referred by her physicians to the Creighton Osteoporosis Center for evaluation of what appeared to be unusually dense bones. She was 18 years old and came to medical attention two years previous because of back pain, which was precipitated by an auto accident in which the car in which she was riding as a passenger was struck from behind. Her only injury was soft tissue injury to her lower back that was manifested by pain and muscle tenderness. There was no evidence of fracture or subluxation on radiographs. The pain lasted for two years, although she was able to attend school full time. By the time she was seen in the Center, the pain was nearly resolved and she was back to her usual activities as a high school student. Physical exam revealed a normal healthy young woman standing 66 inches and weighing 128 pounds. Radiographs of the entire skeleton revealed dense looking bones with thick cortices. All bones of the skeleton were involved. Most importantly, th...
example 2
[0720]The present invention describes DNA sequences derived from two BAC clones from the HBM gene region, as evident in Table 7 below, which is an assembly of these clones. Clone b200e21-h (ATCC No. 980812; SEQ ID NOS: 10-11) was deposited at the American Type Culture Collection (ATCC), 10801 University Blvd., Manassas, Va. 20110-2209 U.S.A., on Dec. 30, 1997. Clone b527d12-h (ATCC No. 980720; SEQ ID NOS: 5-9) was deposited at the American Type Culture Collection (ATCC), 10801 University Blvd:, Manassas, Va. 20110-2209 U.S.A., on Oct. 2, 1998. These sequences are unique reagents that can be used by one skilled in the art to identify DNA probes for the Zmax1 gene, PCR primers to amplify the gene, nucleotide polymorphisms in the Zmax1 gene, or regulatory elements of the Zmax1 gene.
example 3
Yeast-2 Hybrid Screen for Peptide Aptamer Sequences to Dkk-1
[0721]Peptide aptamer library construction. A peptide aptamer library, Tpep, was constructed, which provides a means to identify chimeric proteins that bind to a protein target (or bait) of interest using classic yeast two hybrid (Y2H) assays. The Tpep library is a combinatorial aptamer library composed of constrained random peptides, expressed within the context of the disulfide loop of E. coli thioredoxin (trxA), and as C-termini fusion to the S. cerevisiae Gal4 activation domain. The Tpep library was generated using a restriction enzyme modified recombinant Y2H prey vector, pPC86 (Gibco), which contains the trxA scaffold protein.
[0722]Generation of aptamer-encoding sequences. Aptamer-encoding sequences were produced as follows. DNA encoding random stretches of approximately sixteen amino acids surrounded by appropriate restriction sites were generated by semi-random oligonucleotide synthesis. The synthetic oligonucleotid...
PUM
| Property | Measurement | Unit |
|---|---|---|
| bone mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com