Method for determination of the length of the g-tail sequence and kit for the method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Test for Confirming Dose-Response of Synthetic Single-Stranded G Tail
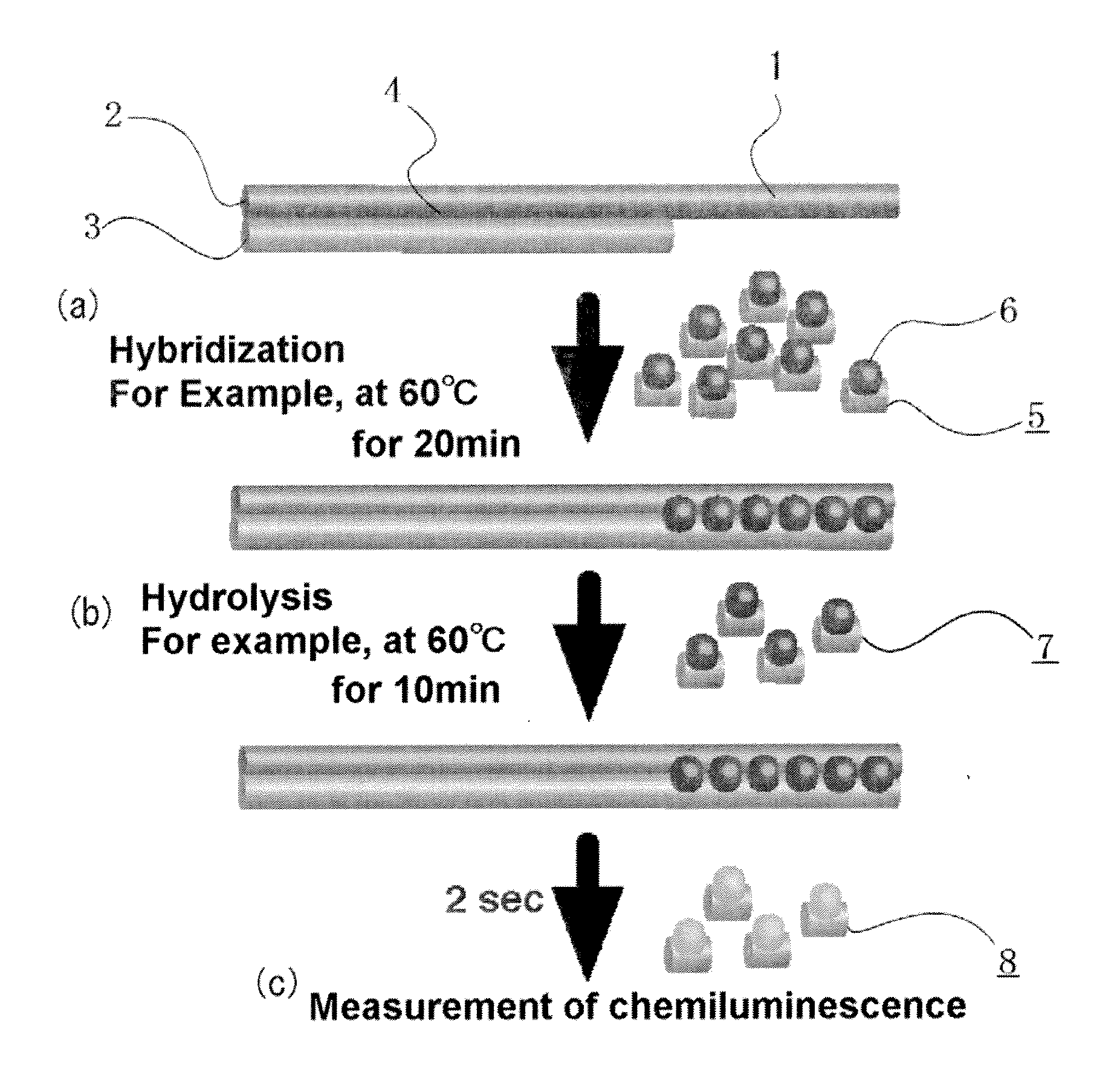
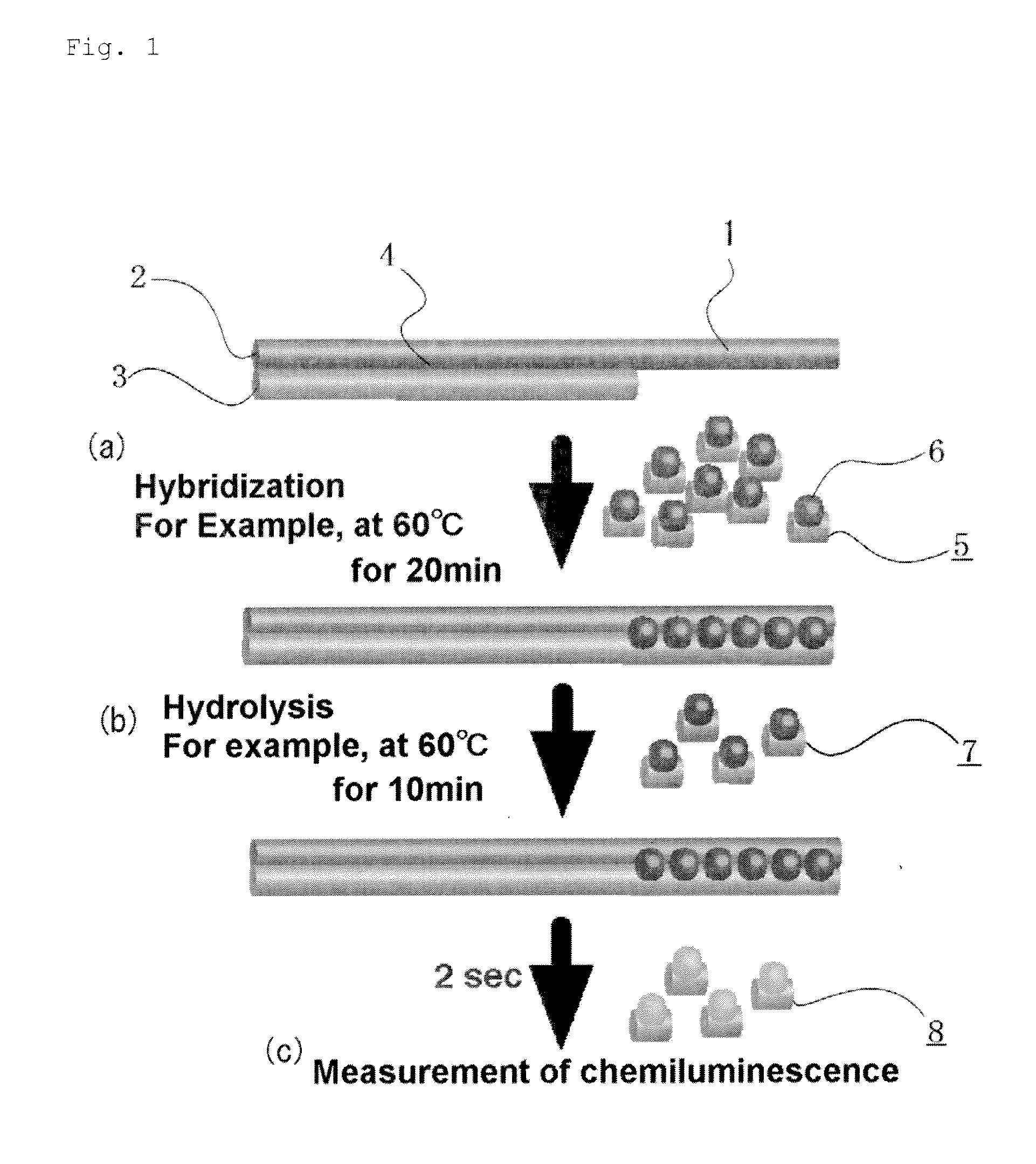
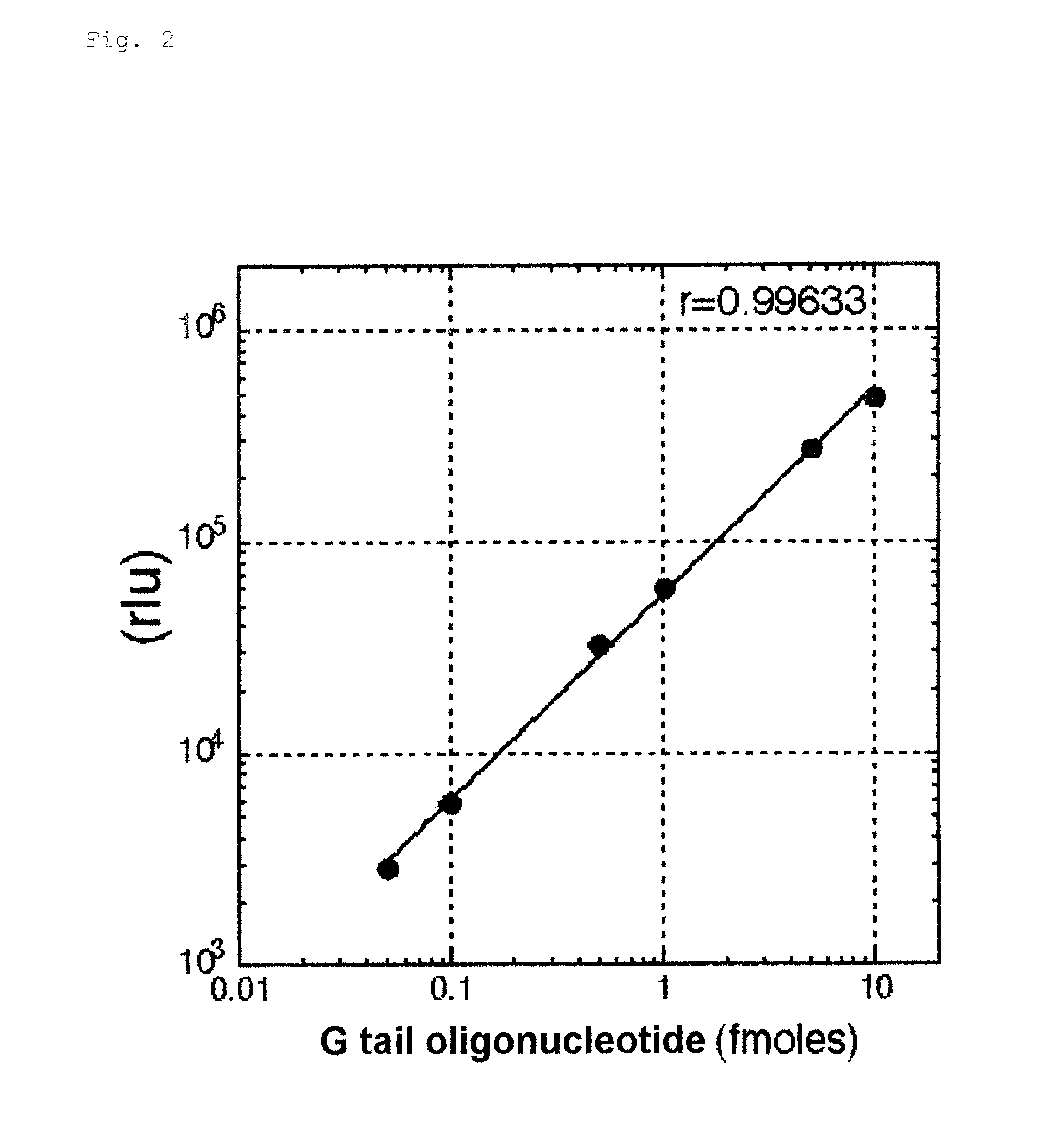
[0125]For the confirmation of dose-response relationship by the measuring method according to the invention, the following hybridization buffer diluents containing a 84-base single-stranded synthetic G tail, 5′-(TTAGGG)14-3′ (manufactured by Prorigo) at various concentrations and an AE-labeled G tail HPA probe (5′-CCCTAACCCTAACC*CTAACCCTAACCCTA-3′, SEQ ID No. 1, *: AE-labeling site, 29 bases) having a amount of chemiluminescence of 3×107 relative light units (hereinafter, referred to simply as rlu) were incubated and allowed to hybridize with each other in 100 μL of the following hybridization buffer at 60° C. for 20 minutes.
[0126]The AE-labeled G tail probe was prepared by labeling, with AE, an amino linker-introduced oligonucleotide (SEQ ID No. 1) prepared by using the linker-introducing reagent 3, according to the method described in Japanese Patent No. 3483829.
TABLE 2Composition of hybridization buffer0.1 mol / ...
example 2
Measurement of G tail sequence length, directly by using cell pellet (1)
[0152]The SiHa cancer cell line pellet prepared in the section below for the measurement of the length of the G tail in cell pellet was resuspended in 100 μL of the hybridization buffer above, and the suspended solution was mixed by pipetting and sheared with a 26G syringe before use. The AE-labeled G tail HPA probe in an amount of 3×106 rlu and the cell pellet were incubated and hybridized under a condition similar to that in above. Hydrolysis and chemiluminescence detection of the AE in the unhybridized probe were performed under a condition similar to that in above. The cell pellet (1 / 10 volume) was denatured similarly to above and hybridized with 3×106 rlu of the AE-labeled Alu probe, for normalization of the total genomic DNA amount.
Preparation of Cell Pellet
[0153]A cell pellet of a SiHa cancer cell line was prepared by collecting cells after the centrifugation of the SiHa cancer cell line at 1,000 G ...
example 3
Test for Comparing the Measuring Method According to the Invention with that Described in Japanese Unexamined Patent Publication No. 2001-95586
[0164]The dominant negative allelic gene (TRF2ΔBΔM) was infected to various cells (HeLa cancer cell, SiHa cancer cell, MCF-7 cancer cell, MRC-5-hTERT normal fibroblast, and 90p normal mammary epithelial cell) according to a method similar to that in ; The nondenatured DNA was isolated from each cell (HeLa cancer cell, SiHa cancer cell, MCF-7 cancer cell, MRC-5-hTERT normal fibroblast, and 90p normal mammary epithelial cell) according to a method similar to that in ; The nondenatured DNA (5 μg) was used in the G tail measuring method according to the present invention for measurement of the G tail length; and, as a Comparative Example, the measuring method described in Japanese Unexamined Patent Publication No. 2001-95586 was applied to each denatured DNA (0.5 μg) for measurement of the entire telomere length. ExoI treatment was performed sim...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com