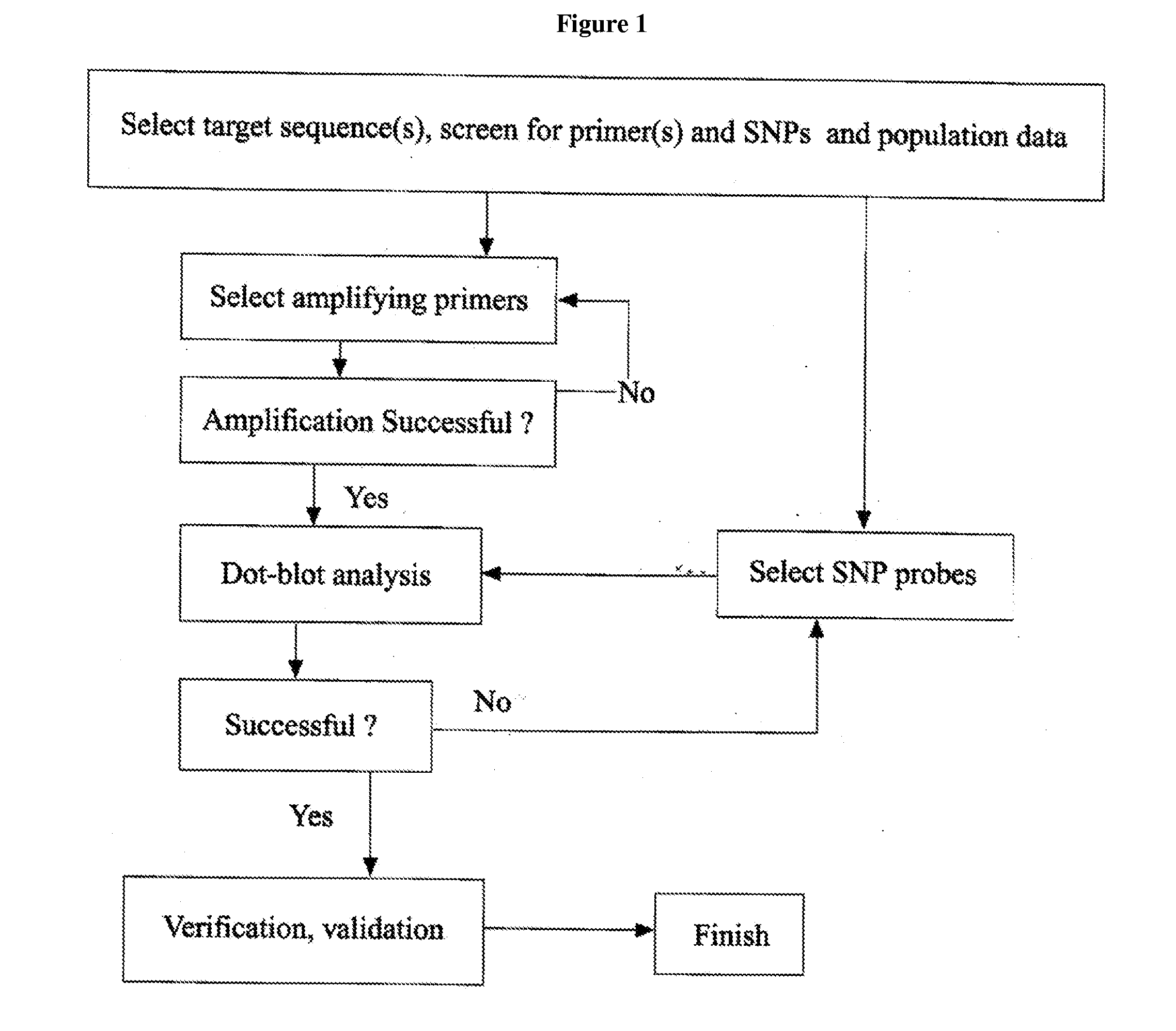
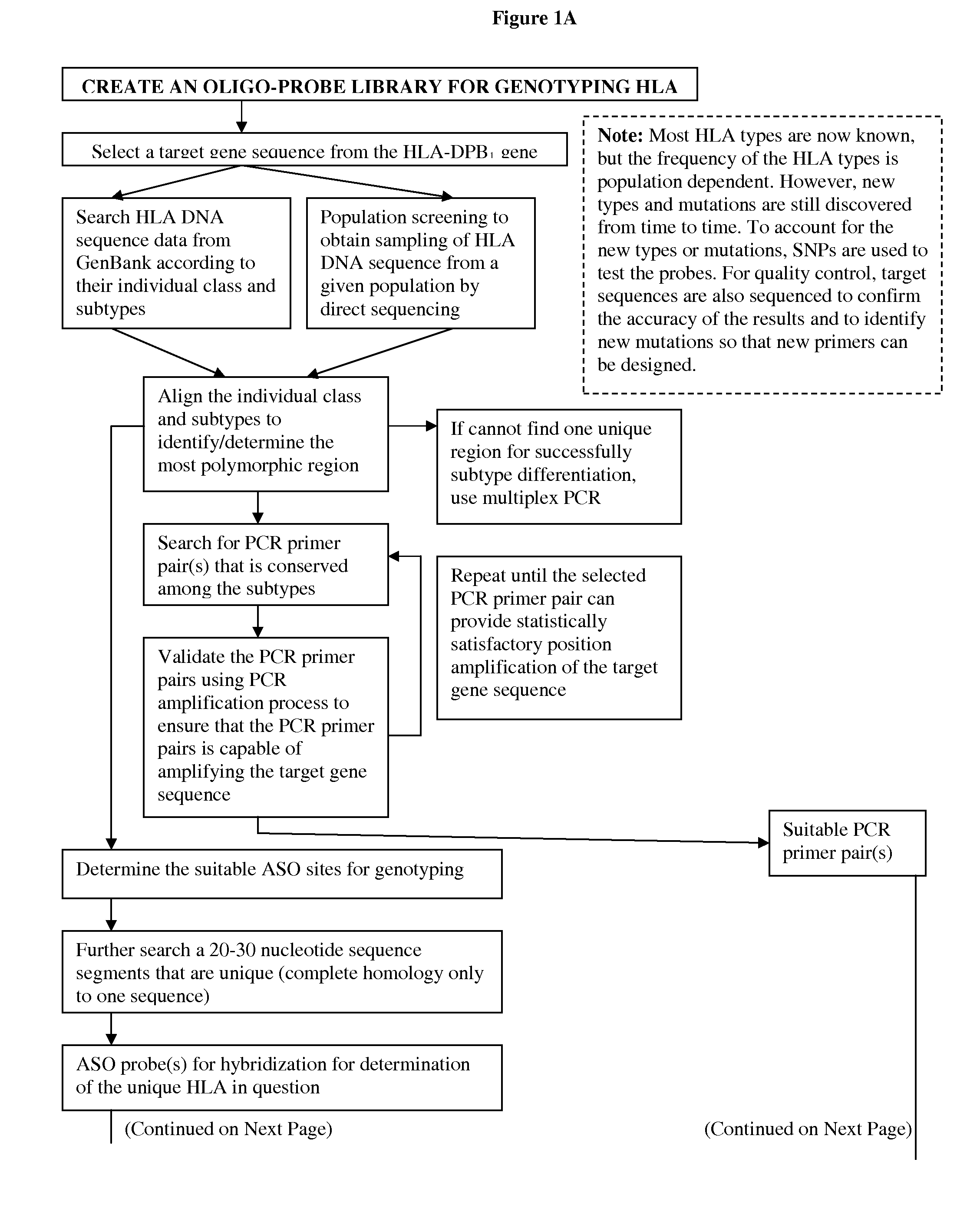
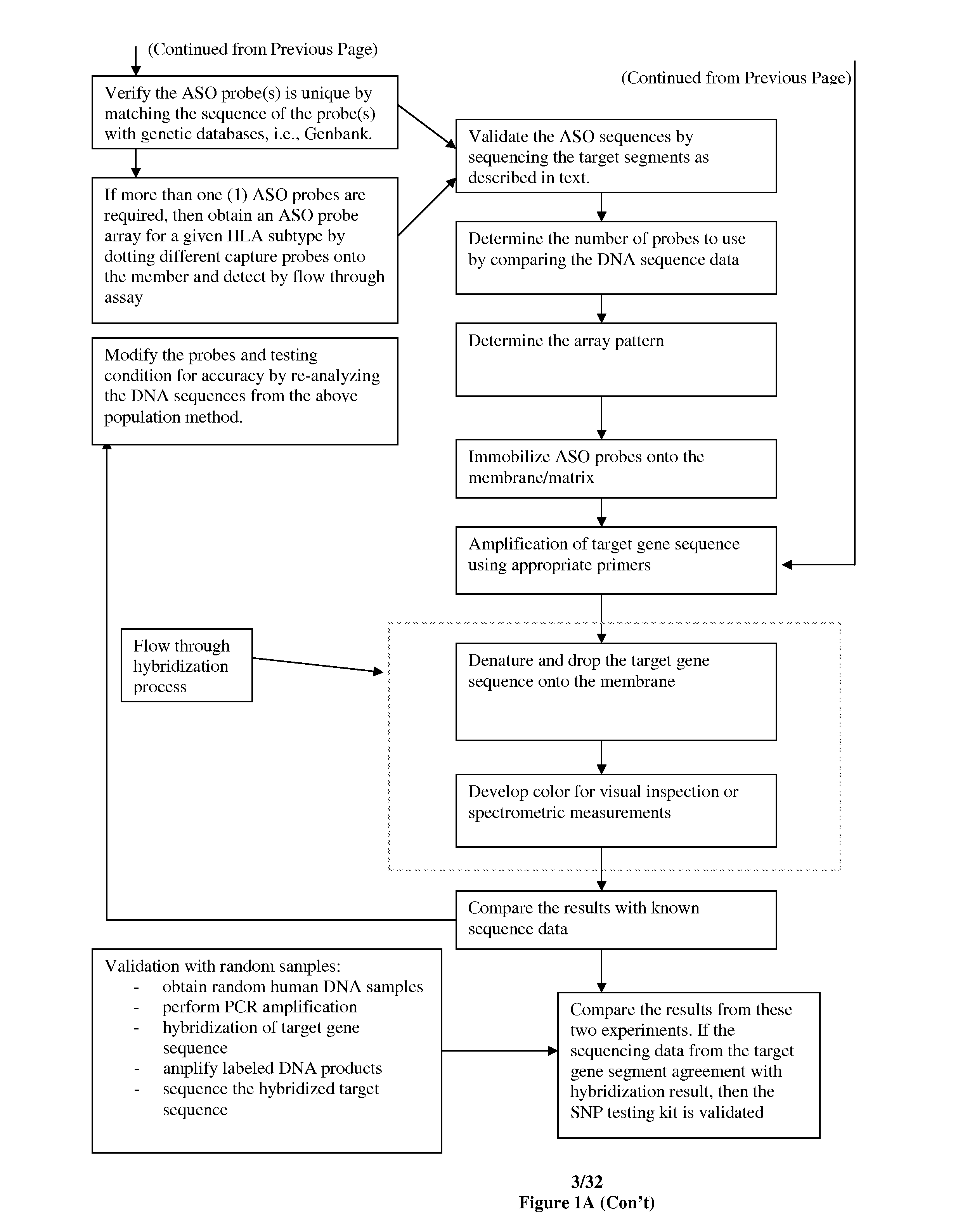
Rapid genotyping analysis for human papillomavirus and the device thereof
a technology of human papillomavirus and genotyping analysis, which is applied in the field of human papillomavirus genotyping analysis, can solve the problems of inability to guarantee the results of hla classification, frequent uninterpretable sequencing data, and direct sequencing, etc., and achieves the effects of accurate genotyping determination, cost saving, and simple and fast technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of DNA
[0087]The following protocols are recommended, but alternative procedures which are readily apparent to one of ordinary skill in the art and which are equally effective may be employed. Nucleated cells such as white blood cells or tissues are washed with PBS, centrifuged, and the supernatant is removed. The pellet is re-suspended in 200 μl PBS. DNA extraction is performed with QIAamp DNA mini kit (QIAGEN) following Blood and Body Fluid Spin protocol as recommended by the manufacturer. Other commercially available kits for isolating DNA may also be used. However, DNA isolation procedures that produce purified DNA that do not contain DNA polymerase inhibitor are crucial to ensure efficient amplification. Elute DNA in 50-200 μl Buffer AE and store at −20° C. until use.
PCR Amplification
[0088]Since PCR is extremely sensitive process, special care must be taken to prevent cross contamination and / or to prevent false positive results. Hence, the following guid...
example 2
Simplified Genotyping Protocols and Devices
[0115]The flow-through DNA hybridization method and device as described in the U.S. Pat. Nos. 5,741,647 and 6,020,187, respectively, reduces hybridization time from many hours or days to minutes (the whole hybridization assay can be completed in 5-30 minutes depending the method used to generate detection signal). The device is also inexpensive to manufacture, and uses 10 times less reagents than convention hybridization devices which will lead to more affordable DNA diagnosis technology.
[0116]The present invention provides an inexpensive platform for studying the nucleic acids, proteins and other chemical interactions using a low-density array format. As illustrated above, the genotyping method of the present invention has been shown to provide significant improvements over conventional hybridization processes.
[0117]The present invention further provides additional improvements over existing flow-through hybridization techniques. For examp...
example 3
Single Nucleotide Polymorphism (SNP)-Based Genotyping
[0122]Eight gene clusters and 55 segments from 50 to 400 individual samples were sequenced to identify sites suitable for SNP genotyping. FIG. 9 shows one of the panels which were used for fingerprinting. Results were compared with STR Profiler Plus fingerprinting kit (Applied Biosystems, Inc.) to ensure accuracy. FIG. 10 shows the loci used in the fingerprinting method as shown in FIG. 8. Other probes and primers for other candidate genes / sequences may be readily determined by one of ordinary skill in the art following the teaching of this application. Genes which have been tested include Globin genes for Thalassemia, BRCAs, ApoE, Collagens, p53, G6PD deficiency alleles and HLA DP, DQ and DR. Any known SNPs of any organisms with adequate data to perform genetic analysis can be tested or detected using the rapid SNP genotyping process of the present invention.
[0123]To identify the DRB genotypes, the PCR were carried out with the p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| gel electrophoresis size | aaaaa | aaaaa |
| restriction fragment length polymorphism | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com