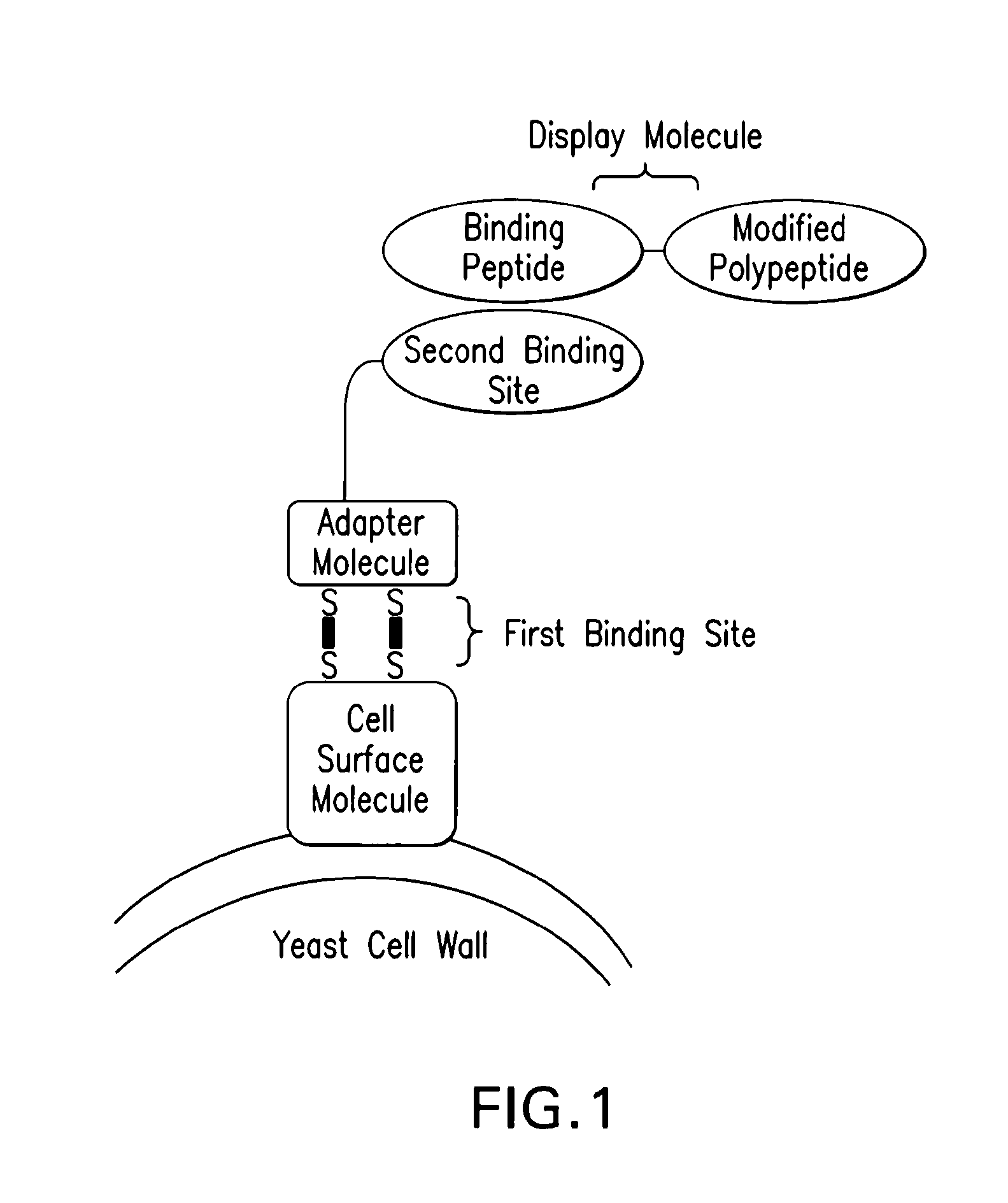
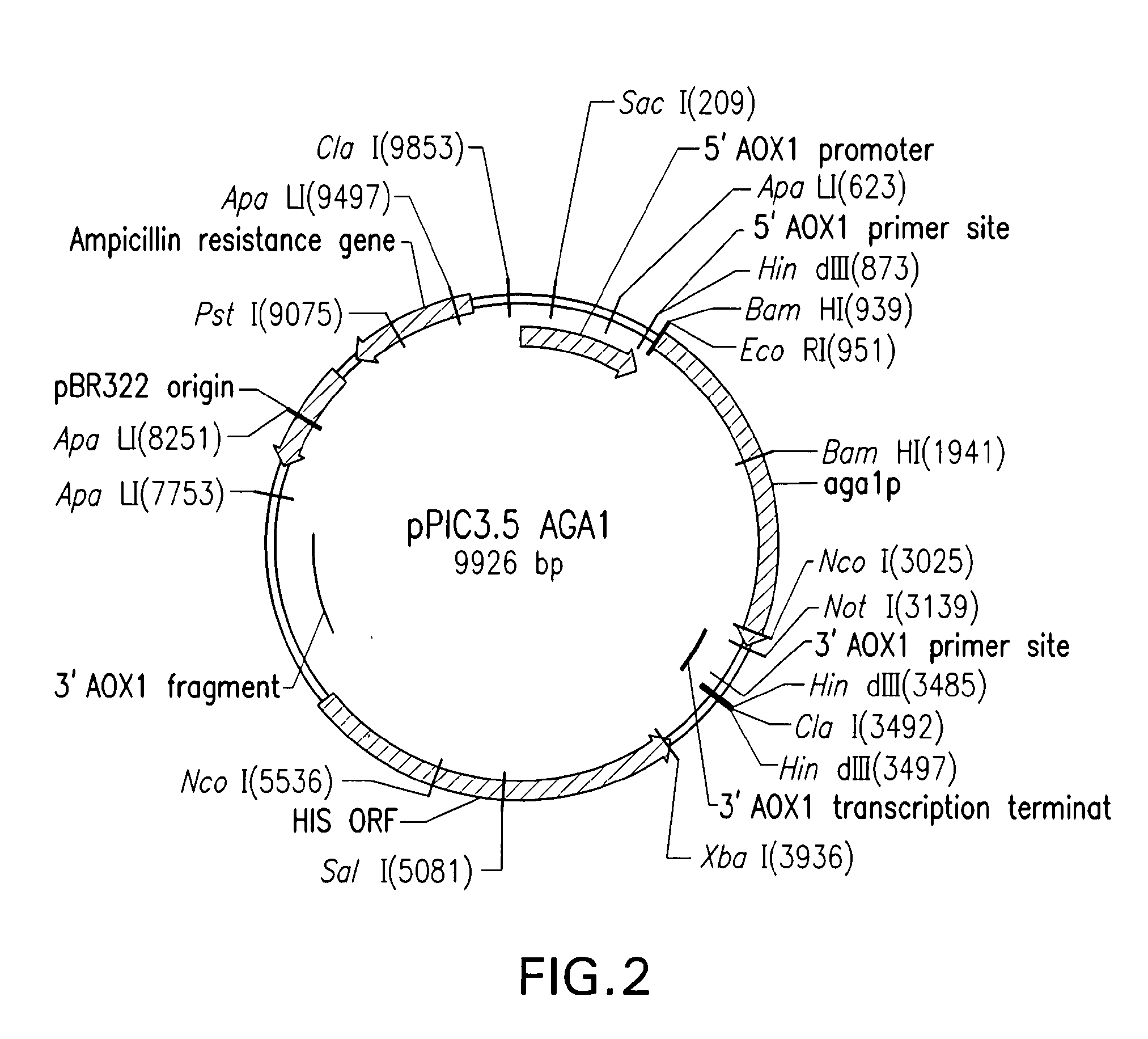
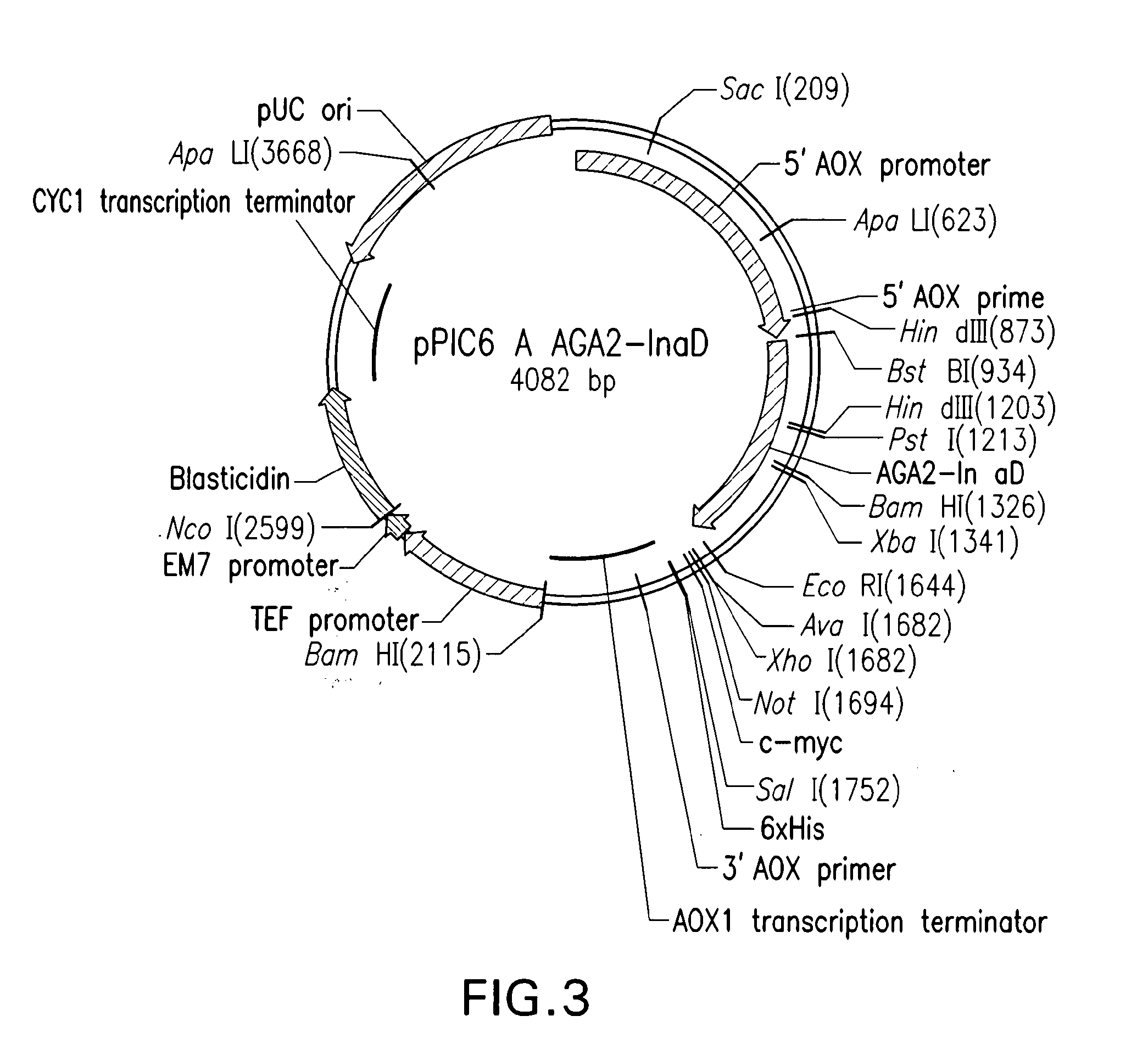
Yeast Display Systems
a display system and protein technology, applied in the field of protein display libraries and library screening, can solve the problems of difficult to determine the actual affinity of a target protein for the phage display protein, require post-translational modifications, and not always identify the target protein that binds the library with very high affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Yeast Display Using InaD / NorpA Interaction in Pichia pastoris
The protein display system for P. pastoris was developed to display a fibronectin type 111 domain (Fn10), by fusing a hybrid secretion sequence (MFalpha / HSA) or a yeast leader sequence (MFalpha1) at N-terminus of Fn10 and fusing the NorpA ligand to its C-terminus. Once expressed, the Fn10 was secreted from the cell and the NorpA ligand bound specifically to the PDZ domain of InaD through disulfide bonds. The InaD was fused to the C-terminus of the Agap2 protein. The Aga2p-InaD fusion protein served as the adapter protein, and the N-terminal Aga2p bound Aga1p, which was immobilized on the surface of the cell. Aga2p bound specifically to Aga1p through disulfide bonds.
The three component system consisting of the Fn10-NorpA fusion protein, the
Aga2p-InaD fusion protein, and the Aga1p cell surface protein were cloned into pPIC expression vectors, under the control of an inducible promoter. The inducible promoter used was the A...
example 2
Switch System to Secrete or Display Fibronectins on the Surface of Pichia pastoris
One variant of the above display system enables the choice between secretion and display of proteins from P. pastoris. To achieve this, the fibronectin construct consisting of the MFalpha1 / HSA hybrid leader followed by fibronectin fused at the C-terminus to the NorpA ligand sequence is cloned into pPlCHOLl-C instead of pPlCHOLl-1. The resulting vector is named pPlCHOLl-C Mfalpha1Hsa-Fn10-NorpA (FIG. 8). The key difference between the two vectors is the promoter, which in pPlCHOLl-1 is the AOX1 promoter induced by methanol, and in the pPlCHOLl-C is the Cup1 promoter induced by copper. To display the fibronectin on the surface of P. pastoris, AGA1 and AGA2-InaD are induced with methanol, while pPlCHOL1-C is induced with copper. This allows for the capture of the secreted fibronectin on the surface of yeast mediated through the tight InaD / NorpA interaction. For secretion of the fibronectin without displa...
example 3
Yeast Display Using InaD / NorpA Interaction in Saccharomyces cerevisiae
This example describes using the InaD / NorpA system with other yeast strains such as Saccharomyces cerevisiae
Strains and Media
Escherichia coil Top10 (Invitrogen, Carlsbad, Calif.) was used as the host strain for recombinant DNA manipulation. The S. cerevisiae strain EBY100 (Invitrogen Co., Carlsbad, Calif.) was used for the production of the fusion proteins AGA2-InaD and MSalpha1 / HSA-Fn10-NorpA or pYS6CT*MFalpha1-HSA-NorpA. E. coli was cultivated LB medium (1% tryptone, 0.5% yeast extract, and 0.5% sodium chloride) containing 100 u / g / mL ampicillin or 100 ug / ml Blasticidin. EBY 100 was cultivated in CM medium-URA.
Construction of Expression Plasmids
The InaD anchor gene was synthesized by Geneart (Germany) and subcloned in frame with the AGA2 anchor protein into the expression vector pYD NBC1 (derivative of pYD1Invitrogen) using HindIII and EcoRI restriction sites. The resulting vector was named pYD_NBC1 AGA2-InaD (...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| MW | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com